ABSTRACT
Pythium species are ubiquitous organisms known to be pathogens to terrestrial plants and marine algae. While several Pythium species (hereafter, Pythium) are described as pathogens to marine red algae, little is known about the pathogenicity of Pythium on marine green algae. A strain of a Pythium was isolated from a taxonomically unresolved filamentous Ulva collected in an intertidal area of Oslo fjord. Its pathogenicity to a euryhaline Ulva intestinalis collected in the same area was subsequently tested under salinities of 0, 15, and 30 parts per thousand (ppt). The Pythium isolate readily infected U. intestinalis and decimated the filaments at 0 ppt. Mycelium survived on U. intestinalis filaments for at least 2 weeks at 15 and 30 ppt, but the infection did not progress. Sporulation was not observed in the infected algal filaments at any salinity. Conversely, Pythium sporulated on infected grass pieces at 0, 15, and 30 ppt. High salinity retarded sporulation, but did not prevent it. Our Pythium isolate produced filamentous non-inflated sporangia. The sexual stage was never observed and phylogenetic analysis using internal transcribed spacer suggest this isolate belongs to the clade B2. We conclude that the Pythium found in the Oslo fjord was a pathogen of U. intestinalis under low salinity.
AbbreviationsCMA P5ARP
cornmeal agar amended with 5 mg L−1 pimaricin, 250 mg L−1 ampicillin, 10 mg L−1 rifampicin, and 100 mg L−1 pentachloronitrobenzene COI
cytochrome coxidase subunit I HSD
honestly significant difference ITS
internal transcribed spacer MQ
Milli Q PCA
potato carrot agar Por
Porphyra Pyr
Pyropia Pyt
Pythium ppt
parts per thousand rbcL
ribulose bisphosphate carboxylase large subunit V8A
eight vegetables juice agar INTRODUCTION
Pythium Pringsheim is a genus of parasitic and saprophytic oomycetes, which form a distinct phylogenetic lineage of fungus-like eukaryotes. They are filamentous and microscopic and reproduce both sexually and asexually. Oomycetes are placed under the kingdom Chromista, which includes a diverse groups of photosynthetic organisms such as the large brown seaweeds and microscopic diatoms (Cavalier-Smith 2018).
Pythium species are ubiquitous organisms found in aquatic and terrestrial habitats from the tropics to the Polar regions. Pythium species are best known as plant pathogens (Hendrix and Campbell 1973), but they have also been described as pathogens of crabs and fish (Czeczuga 1996), mammals (Gaastra et al. 2010), and algae (Van der Plaats-Niterink 1981). They play an important role as debris decomposers and they have been isolated from dead insects, decaying plant material, water and soil (Van der Plaats-Niterink 1981).
The genus Pythium was divided in 11 clades (from A to K) based on the internal transcribed spacer (ITS) region and the D1 to D3 regions of the nuclear large ribosomal subunit (Lévesque and De Cock 2004). One of the clades, clade K, was later segregated from the genus and described as the new genus Phytopythium (Bala et al. 2010). Furthermore, Uzuhashi et al. (2010) proposed to segregate the genus Pythium into five different genera.
The first Pythium species described infecting a marine alga was Pyt. marinum Sparrow found on red algae Ceramium rubrum (= Ceramium virgatum Roth) in Denmark (Sparrow 1934). A Pythium sp. causing disease in the cultivated red alga Porphyra tenera [= Pyropia tenera (Kjellman) N. Kikuchi, M. Miyata, M. S. Hwang & H. G. Choi] (nori) was reported from Japan in 1947 (Arasaki 1947), and this was later described as Pyt. porphyrae Takahashi and Sasaki (Takahashi et al. 1977). Pyt. marinum has been reported from North America and Europe mostly infecting Porphyra (= Pyropia) species (Kazama and Fuller 1977, Aleem 1980, Kerwin et al. 1992). Pyt. porphyrae is reported as the causal agent of the red rot disease causing substantial economical loses in nori production in Asia (Kim et al. 2014), and it has been also isolated from natural populations of Pyropia plicata W. A. Nelson in New Zealand (Diehl et al. 2017) and Pyr. acanthophora (E. C. Oliveira & Coll) M. C. Oliveira, D. Milstein & E. C. Oliveira in the Philippines (Dumilag 2019).
There are other Pythium species found in marine environments: Pyt. grandisporangium Fell and Master (Fell and Master 1975, De Cock 1986, Kurokawa and Tojo 2010), Pyt. chondricola De Cock (De Cock 1986, Lee et al. 2015), Pyt. aquatile Höhnk, Pyt. salinum Höhnk (Van der Plaats-Niterink 1981), and Pyt. undulatum H. E. Petersen (Thompson 1982). Several of them have been isolated from algae. For example, Pyt. grandisporangium and Pyt. undulatum were isolated from the brown alga Fucus distichus Linnaeus while Pyt. chondricola was isolated from several decaying red algae such as Chondrus crispus Stackhouse, Pyr. yezoensis (Ueda) M. S. Hwang & H. G. Choi and an unknown species, and from a decaying green alga Ulva lactuca Linnaeus (De Cock 1986, Lee et al. 2015).
The morphology of Pyt. marimum, Pyt. porphyrae, and Pyt. chondricola is very similar and there is controversy about their separation into three species (Park et al. 2000, Lévesque and De Cock 2004). The utilization of molecular methods in the identification of Pythium has not completely resolved the issue. Sequences of the ITS region, currently used in oomycetes taxonomy, are the same for Pyt. porphyrae, Pyt. chondricola, and Pyt. marinum isolated from marine habitats (Park et al. 2001, Lévesque and De Cock 2004). On the other hand, when comparing cytochrome c oxidase subunit I (COI) sequences, it was possible to discriminate among Pyt. porphyrae and Pyt. chondricola (Robideau et al. 2011, Lee et al. 2015). Recent studies further showed that cytochrome c oxidase subunit 2 sequences can also distinguish between these two species (Lee et al. 2017).
Several Pythium species, e.g., Pyt. marinum, Pyt. porphyrae, and Pyt. chondricola, are pathogenic to the gametophytic life stage of Porphyra and Pyropia species (Kazama and Fuller 1977, Kerwin et al. 1992, Diehl et al. 2017, Lee et al. 2017), but were unable to infect the conchocelis life stage of these species (Kerwin et al. 1992, Diehl et al. 2017). Artificial infections of U. lactuca with Pyt. marinum and of U. fasciata Delile, U. pertusa (= U. australis Areschoug), U. prolifera O. F. Müller with Pyt. porphyrae in half-strength seawater have been unsuccessful (Kerwin et al. 1992, Uppalapati and Fujita 2000). Artificial infection of Ulva sp. with Pyt. porphyrae in 1/4 strength seawater has also been unsuccessful (Diehl et al. 2017).
Other Pythium species, e.g., Pyt. adhaerens Sparrow, Pyt. angustatum Sparrow, Pyt. diclinum Tokunaga (syn. Pyt. gracile Schenk sensu Middleton), Pyt. mamillatum Meurs and Pyt. tenue Gobi are pathogenic to freshwater green algae (Van der Plaats-Niterink 1981). However, not much is known about pathogenicity of different Pythium species on marine green seaweeds. Previously, Pyt. chondricola was found on U. lactuca (De Cock 1986), but its pathogenicity was not reported.
Here we present the morphological and molecular characteristics of a Pythium strain isolated from an Ulva species collected from Oslo fjord. We also investigated the potential of the isolated Pythium strain to grow and sporulate and its pathogenicity at different salinities on another Ulva species (i.e., U. intestinalis Linnaeus) collected from the same area.
MATERIALS AND METHODSUlva collectionFilaments of euryhaline Ulva species were collected in an intertidal area of Oslo fjord (59°43′18.2″ N, 10°43′39.1″ E) in August and October 2013. On site, the algae were exposed to extreme salinity fluctuation due to persistent freshwater outflow from a nearby eutrophic stream (Årungselva), seasonal rainfall, and tidal exchange. Fjord seawater salinity was 29 parts per thousand (ppt).
Isolation of Pythium
Ulva filaments collected in August 2013 were examined for the presence of oomycetes. Single threads were carefully separated and immersed in Petri dishes containing sterile Milli-Q (MQ) water. After 4 days, aseptate hyphae with appressoria formation were observed on one of the filaments. Part of the filament was stained with cotton blue for better observation of the hyphal growth (Fig. 1A). The other part of the filament was plated onto semi-selective medium for oomycetes CMA P5ARP (cornmeal agar amended with 5 mg L−1 pimaricin, 250 mg L−1 ampicillin, 10 mg L−1 rifampicin, and 100 mg L−1 pentachloronitrobenzene) (Ferguson and Jeffers 1999). The plate was incubated at room temperature and mycelial growth was transferred onto V8 juice agar (V8A), containing 200 mL V8 juice (Campbell Soup Co., Camden, NJ, USA), 800 mL MQ H2O, 3 g CaCO3, and 15 g agar. The isolate was maintained on pieces of V8A submerged in vials with MQ water at 5 and 15°C. The isolate was deposited in the culture collection of NIBIO at Ås, Norway, with accession No. 231095 and in the Westerdijk Fungal Biodiversity Institute collection, Netherland, with accession No. CBS 139622.
Molecular identification of Ulva spp. and Pythium spTotal genomic DNA was extracted from Pythium mycelia grown on V8A or from algal filaments using the Qiagen DNeasy Plant Mini Kit (Hilden, Germany), and DNA segments were amplified by PCR using Taq polymerase and designated primers. For Pythium, the ITS was amplified and sequenced in both directions using primers ITS5 and ITS4 (White et al. 1990), and part of the COI gene using the oomycete-specific primers OomCoxILevup and Fm85mod (Robideau et al. 2011). A fragment of the ribulose-bisphosphate carboxylase large subunit (rbcL) gene was targeted using primers rbcLa-F and rbcLa-R for the Ulva samples (Kuzmina et al. 2012). The amplicons were sequenced by GATC Biotech, Germany, and the raw data were assembled and analyzed using the Lasergene Sequence Analysis Software (DNASTAR Inc., Madison, WI, USA). The program BLASTN (National Center for Biotechnology Information, Bethesda, MD, USA) was used to search existing DNA databases for sequences showing similarity to the obtained sequences for the three genes. The best hits obtained from the blast searches from the sequences of Pythium and Ulva were downloaded and used in the phylogenetic analyses. We also included representative sequences from Pythium specimens (Robideau et al. 2011, Uzuhashi et al. 2015). In the case of Ulva, representative sequences of Ulva were selected from a previous analysis of Kang et al. (2014). The sequences for the three gene sets (ITS, COI, and rbcL) were aligned and manually inspected in AliView (Larsson 2014). Phylogenetic relationships were inferred on each separate aligned matrix with maximum likelihood as implemented in RAxML ver. 8.2 (Stamatakis 2014) using the GTRCAT model, 1,000 bootstrap and default settings. Pythium undulatum and Umbraulva olivascens (= Umbraulva dangeardii M. J. Wynne & G. Furnari) were used as outgroups for the phylogenetic analyses. Obtained trees were edited with FigTree (Rambaut and Drummond 2016). All new sequences generated in this study are deposited in the GenBank database under accession Nos. MK644144, MK644145, MK644146, and MK644147.
Growth of Pythium and determination of cardinal temperaturesThe Pythium isolate was subsequently grown on V8A and on potato carrot agar (PCA) (Van der Plaats-Niterink 1981) for morphological observations and growth studies. The maximum, minimum, and optimum temperatures for growth as well as the radial growth per day at different temperatures were determined on PCA as described by De Cock and Lévesque (2004) with small modifications. Agar plates were inoculated in the center and placed at room temperature. After 24 h, the plates were incubated at 1, 5, 10, 15, 20, 25, 30, 35, 37, and 40°C. Two PCA plates were used for each temperature. Radial growth was measured at two perpendicular diameters every 24 h for 3 days. After 3 days, the plates without mycelial growth were placed at room temperature for 1 week to check if growth resumed.
Growth of Pythium at different salinitiesCMA plates were prepared by using sterile MQ water or saline solutions at 5, 10, 15, 20, 25, 30, and 35 ppt. The saline solutions were prepared by dissolving Sea Salt (Sigma, St. Louis, MO, USA) in sterile MQ water. The plates were inoculated at the border and incubated at 20°C in the dark. The radial growth along a dish diameter was measured every 24 h for 4 days. Four Petri dishes were use per each salinity.
Sporulation of Pythium in water culture under different salinitiesSporulation at different salinities was studied in grass water cultures. The isolated Pythium was grown on PCA plates for 2 days. Thereafter, 1–2 cm pieces of autoclaved lawn grass were placed on top of the growing Pythium colonies. After 24 h, the grass pieces infected with Pythium were placed in 9 mm Petri dishes containing ca. 20 mL of sterile MQ water or saline solutions at 15 and 30 ppt. The saline solutions were prepared by dissolving Sea Salt (Sigma) in sterile MQ water. The plates with the infected grass pieces immersed in different saline concentrations were incubated at room temperature (22–23°C). The presence or absence of vesicles and zoospores was checked after 2 and 5 h. Observations were made with an invert microscope that allows a quick check of the dishes without disturbance of the samples or the need of slides preparations. The dishes were periodically observed for formation of sexual stages for 2 weeks. The experiment was performed twice.
Pathogenicity assaysThe pathogenicity of the isolated Pythium was tested on a filamentous Ulva species collected in October 2013. The Ulva filaments were carefully examined to make sure that they were not infected with oomycetes. Two pathogenicity tests were performed. In the first test, Ulva filaments were placed on the growing margin of a Pythium colony on PCA. The day after, the infected filaments were transferred to MQ water and two different saline concentrations (15 and 30 ppt). Non-inoculated filaments were placed in MQ water as controls. The filaments were periodically observed for 2 weeks, using an inverted microscope, in order to detect mycelium development and sporulation. To obtain photos, slide preparations were made for use in an upright microscope. After 2 weeks the filaments were transferred onto CMA P5ARP to check the survival of the Pythium sp. at the different saline concentrations. In the second test, the algal filaments were infected with zoospores. Small Petri dishes (5 cm diameter) were perforated with small orifices in the angle formed by the bottom and the wall of the dishes. These dishes were placed inside larger dishes (9 cm diameter) filled with fresh water. Grass pieces inoculated with Pythium, as described above, were placed inside the smaller dish and algal filaments in the larger dishes. There was no direct contact between the algal filaments and the grass pieces. The Pythium infecting the grass pieces produced zoospores that could move from the small dishes to the larger ones though the orifices. The filaments were observed periodically for 2 weeks as in the former assay.
Data handling and statistical analysisAll data except radial growth under different salinities were analyzed qualitatively. Radial growth data were tested for homogeneity of variance (Levene Statistics) and normality (Kolmogorov-Smirnov test). Thereafter, the statistical significance of differences in radial growth as affected by salinity treatment was tested using analysis of variance (ANOVA, p = 0.05). This was followed by Tukey honestly significant difference (HSD) test (p < 0.05) to find means that are significantly different from each other. Statistical analyses were done using the SPSS 11.5 software (SPSS Inc., Chicago, IL, USA).
RESULTSUlva identificationThe Pythium-infected Ulva filaments collected in August 2013 (accession No. MK644145) had a 99.7% sequence identity to the rbcL gene of Ulva linza Linnaeus (accession No. KP233764.1). The Ulva filaments collected in October 2013 (accession No. MK644146) and used in the pathogenicity test had 100% sequence identity to the rbcL gene of several voucher specimens of U. intestinalis. The Ulva sample where Pythium was isolated from belong to the LPP (linza-procera-prolifera) complex (Shimada et al. 2008). Ulva linza and U. prolifera are currently accepted taxonomically while U. procera (K. Ahlner) H. S. Hayden, Blomster, Maggs, P. C. Silva, Stanshope & Waaland is currently regarded as a synonym of U. prolifera (Guiry and Guiry 2019). This group is taxonomically not well resolved and thus the Ulva sample was not conclusively identified. On the other hand, the Ulva used for pathogenicity tests clustered with U. intestinalis samples with a 100% bootstrap values (Fig. 2). Morphologically, U. intestinalis and U. compressa Linnaeus have distinguishing characteristics that separate them from other filamentous Ulva species. Moreover, sequences of rbcL can clearly distinguish these two species (Hayden et al. 2003).
Pythium identificationThe ITS sequence of the isolated Pythium (accession No. MK644147) has 98.9% sequence identity to isolates deposited in GenBank as Pyt. dissotocum (accession Nos. KM061705.1 and KM061706.1); however, these two isolates lack COI sequence in GenBank. The COI sequence of the isolated Pythium (accession No. MK644144) has 97.3% identity to Pyt. brachiatum isolate UZ00746 (KJ995594.1). Our phylogenetic analyses based on ITS and COI sequences suggest that the Norwegian isolate belongs to the clade B2 of Lévesque and De Cock (2004) (Fig. 3).
Growth of Pythium and determination of cardinal temperatures
Pythium sp. colonies showed a chrysanthemum-rosette pattern on V8A and PCA (Fig. 1B). The radial growth per day at different temperatures is shown in Fig. 4. At 35°C, limited growth was observed that ceased after day 1; however, growth resumed when culture was transferred to room temperature. At lower temperature, growth ceased at 1°C, but growth resumed when the culture was transferred to room temperature. The main hypha was up to 7 μm in diameter on PCA. The Pythium sp. produced filamentous sporangia that was not easily distinguishable from hyphae. The sexual stage was not observed neither in PCA, V8A, nor in grass in water cultures.
Pythtium growth at different salinitiesRadial growth at different salinities is shown in Fig. 5. Analysis of variance showed that salinity significantly affected radial growth of Pythium sp. (p < 0.001). Posthoc Tukey’s HSD showed growth was highest at salinity 10 = 15, 10 > 20, 15 = 20 > 25 = 5 > 35 > zero. The sexual stage was not observed at any salinity.
Sporulation at different salinitiesThe Pythium was observed to sporulate in fresh and saline waters, but sporulation was faster in freshwater than in saline water. Already after 2 hours of immersion in fresh water, sporulation was observed in the infected pieces of grass in both performed trials. After 2 hours in salinity 15 ppt, the Pythium sporulated in one of the trials and produced only vesicles in the other. In salinity 30 ppt, no sporulation was observed after 2 h; however, in one trial, vesicle formation was observed after 2 h. After 5 h, zoospore release was observed in all treatments.
Pathogenicity testWhen U. intestinalis filaments were placed in close proximity of the mycelium of a Pythium colony, they were readily infected. The hyphae that came in contact with the seaweed formed a large amount of appressoria on the surface of the filaments (Fig. 1C). When Pythium in Ulva filaments was cultured in fresh water, it grew very well. After 5 days, the Pythium had fully colonized the algal filaments (Fig. 1D), and after 2 weeks, the filaments became brownish and completed surrounded by Pythium hyphae (Fig. 1E). Conversely, when Pythium and Ulva filaments were incubated in salinity 15 and 30 ppt, no Pythium growth was observed, and after 2 weeks these algal filaments remained green. The non-infected Ulva filaments (controls) placed in fresh water also remained green. After 2 weeks, the infected filaments were plated on CMA P5ARP and Pythium was re-isolated from freshwater and the two saline concentrations. The Pythium re-isolated from diseased brown filaments was identified by ITS sequencing, confirming that it was the same Pythium sp. that was used for inoculation. Sporulation was never observed in Ulva filaments during the 2-week assay period, neither in fresh water nor in any of the saline concentrations.
In the second test, zoospores produced by Pythium also readily infected the algal filaments. Chemotaxis was not observed; the zoospores seemed to randomly swim and invade the seaweeds. Appressoria formation and mycelial growth on Ulva intestinalis were observed 1 week after inoculation. After 2 weeks, the algal filaments became brownish and there was abundant mycelial growth around the filaments.
DISCUSSION
Ulva intestinalis has typically been found in fresh and saline waters (Edwards et al. 1988). The different ecotypes are adapted to grow in different salinities and populations inhabiting brackish habitats have a wider range of salinity tolerance (Reed and Russell 1979). The survival of U. intestinalis at 0 ppt seems to be very variable. Edwards et al. (1988) reported that U. intestinalis could survive 6 days at 0 ppt, while Ritchie and Larkum (1985) reported a longer survival period of 3–5 weeks. Conversely, no survival at salinity < 1 ppt was observed in another study (Martins et al. 1999). Survival in zero or low salinities is promoted and compensated by the presence of high nutrient levels (Kamer and Fong 2001, McAvoy and Klug 2005). However, the Ulva filaments thriving in low salinity environments are prone to Pythium infection that is of freshwater origin.
The ITS sequence of Pythium in this study is very similar to the sequences of different Pythium species isolated from freshwater samples in the Falkland Islands (Davis 2016). The two isolates with sequences similar to the Pythium isolate described here, are both deposited as Pyt. dissotocum in GenBank. However, in the work of Davis (2016) these isolates are not morphologically described. The Pythium from Oslo fjord belongs to clade B2 of the Pythium phylogenetic tree of Lévesque and De Cock (2004). The common feature of different species in clade B2 is the production of filamentous sporangia. This type of sporangium corresponds to the Pythium sensu stricto in the classification of Uzuhashi et al. (2010). No other Pythium species isolated from marine algae seemed to belong to clade B. In the study of Lévesque and De Cock (2004) there is a Pyt. marinum placed in clade B2, but the authors pointed out that the isolate do not have a maritime origin and is probably misidentified. Due to the absence of sexual stage, it is not possible to make a complete morphological description of the Pythium isolated from Ulva filaments in this study.
The external mycelium developed profusely only when the filaments were placed in fresh water. Sporulation was never observed from mycelium growing from algae at any salinity tested. Zoospores were very easily produced from infected grass pieces. The increase in salinity slightly retarded the Pythium sporulation on grass but was able to sporulate at a salinity of 30 ppt. The tolerance of Pythium species to salinity is very variable. Pyt. undulatum isolated in estuaries from Fucus distichus could sporulate well between 0 and 15 ppt, but sporulation rarely occurred at 27 ppt (Thompson 1982), while Pyt. marinum infecting Por. perforata had difficulties to sporulate at salinities lower than 24 ppt (Kazama and Fuller 1977). In axenic culture, the Pythium of Oslo fjord could grow in all salinities tested with maximum radial growth at 10–15 ppt. Pyt. porphyrae isolated from Pyropia species could also grow at salinities between 0 and 30 ppt with an optimum at 15 ppt (Klochkova et al. 2017). Pyt. grandisporangium isolates from a saline march in Osaka Bay could grow between 0 and 90 ppt with an optimum between 10 and 30 ppt (Kurokawa and Tojo 2010).
The Pythium species found in Oslo fjord is a pathogen of filamentous Ulva species. The infection does not progress at high salinity, but the Pythium can survive in infected algae for at least 2 weeks at 30 ppt. Ulva species exposed to fluctuating salinities, especially towards the lower range, are very likely more prone to infections caused by this Pythium species. Sporulation on infected algae was never observed, but it could sporulate on grass pieces in fresh water and up to 30 ppt. It is probable that terrestrial plants are hosts of this Pythium and floating terrestrial plant debris from freshwater systems could act as vectors infecting intertidal algae exposed to extreme salinity fluctuations.
ACKNOWLEDGEMENTSThis study was supported by the Norwegian Biodiversity Information Centre (Artsdatabanken), project number 70184222.
Fig. 1
Pythium sp. isolate NIBIO 231095. (A) Hyphae infecting a filament of Ulva collected in an intertidal area of Oslo fjord. Pythium hyphae are stained with cotton blue. (B) Colony pattern on potato carrot agar after 5 days at 20°C in the dark. (C–E) Pathogenicity test. (C) Appressoria formation on the surface of Ulva intestinalis after inoculation. The inoculation was made by putting Ulva filaments in contact with a growing Pythium colony. (D) Mycelial growth on the surface of U. intestinalis 5 days after inoculation and incubation at room temperature (22–23°C) in fresh water. Pythium hyphae are stained with cotton blue. (E) Mycelial growth from U. intestinalis 2 weeks after inoculation at room temperature (22–23°C) in fresh water. (F) Sporulation of Pythium sp. in grass water culture 2 h after immersion of grass pieces in fresh water at room temperature. V, vesicle; Z1, clump of realized zoospores; Z2, realized detached zoospore. Scale bars represent: A, 10 μm; C–F, 20 μm. 
Fig. 2Maximum likelihood tree of two Ulva samples collected in an intertidal area of Oslo fjord inferred from rbcL sequences. Numbers at nodes indicates bootstrap support values. GenBank accession numbers are given after each sample in the tree. Scale bar represents substitution rates. 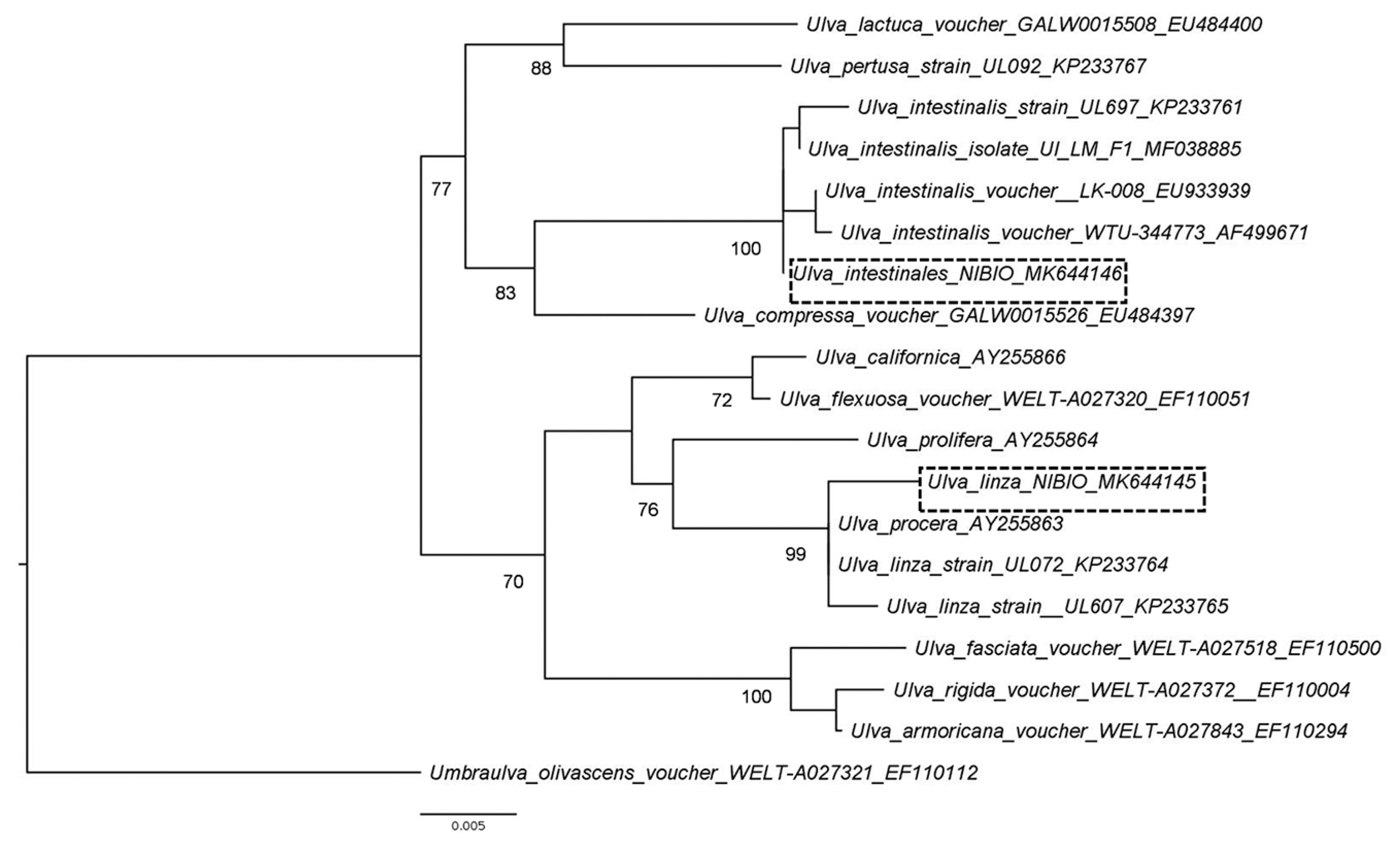
Fig. 3Maximum likelihood tree of Pythium sp. isolate NIBIO 231095 inferred from internal transcribed spacer sequences (A) and cytochrome c oxidase subunit I sequences (B). The sampling includes closely related species belonging to Pythium clade B2 (Lévesque and De Cock 2004, Uzuhashi et al. 2015) and species from other clades with filamentous non-inflated sporangia or found in marine environments. Numbers at nodes indicates bootstrap support values above 80, except the location of the NIBIO isolate in B. GenBank accession numbers are given after each species in the tree. Scale bar represents substitution rates. 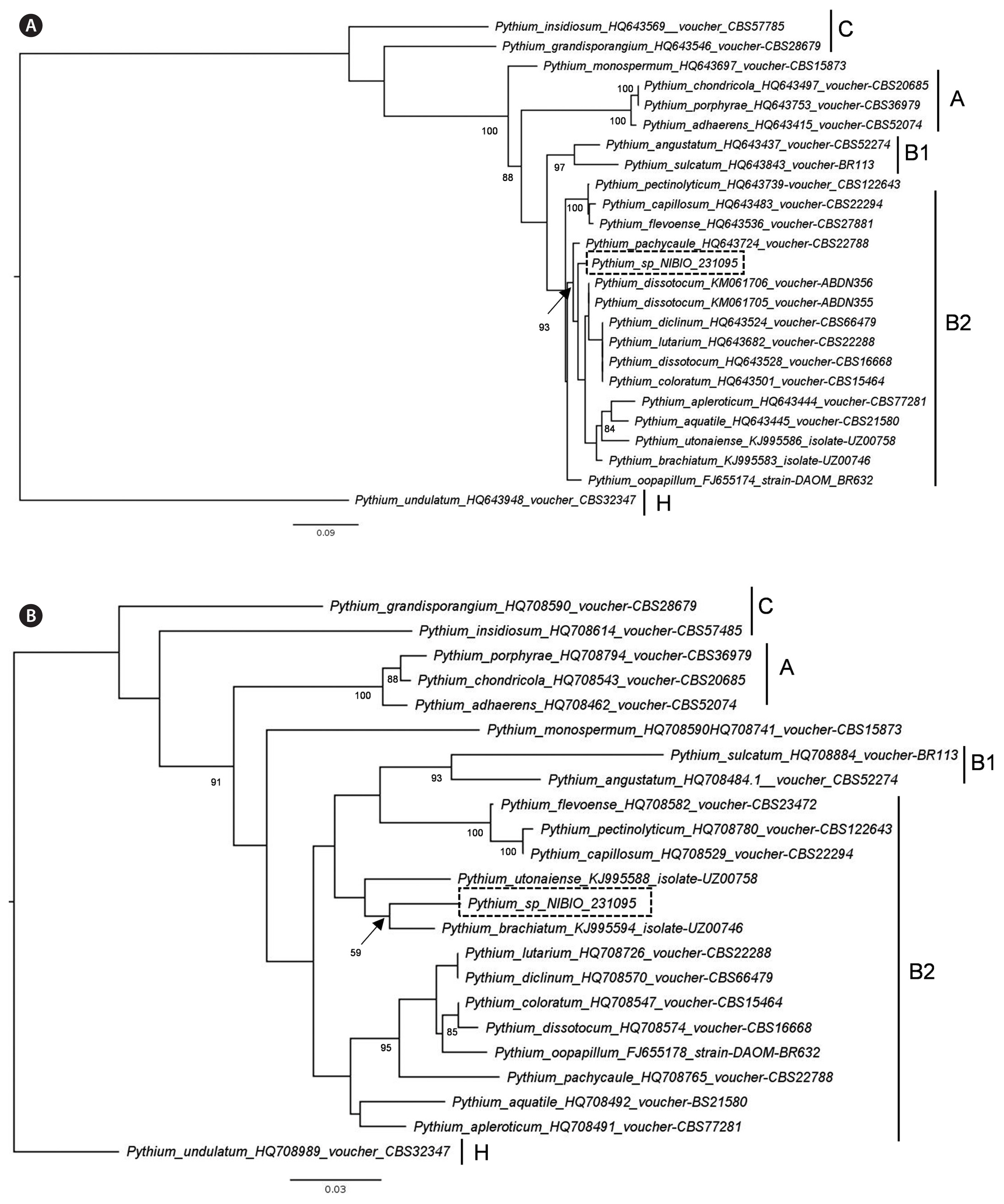
Fig. 4Radial growth (mm d−1) of Pythium sp. isolate NIBIO 231095 on potato carrot agar (0 ppt salinity) at different temperatures. Values are means (± standard deviation, n = 2), data was not subjected to statistical analysis. 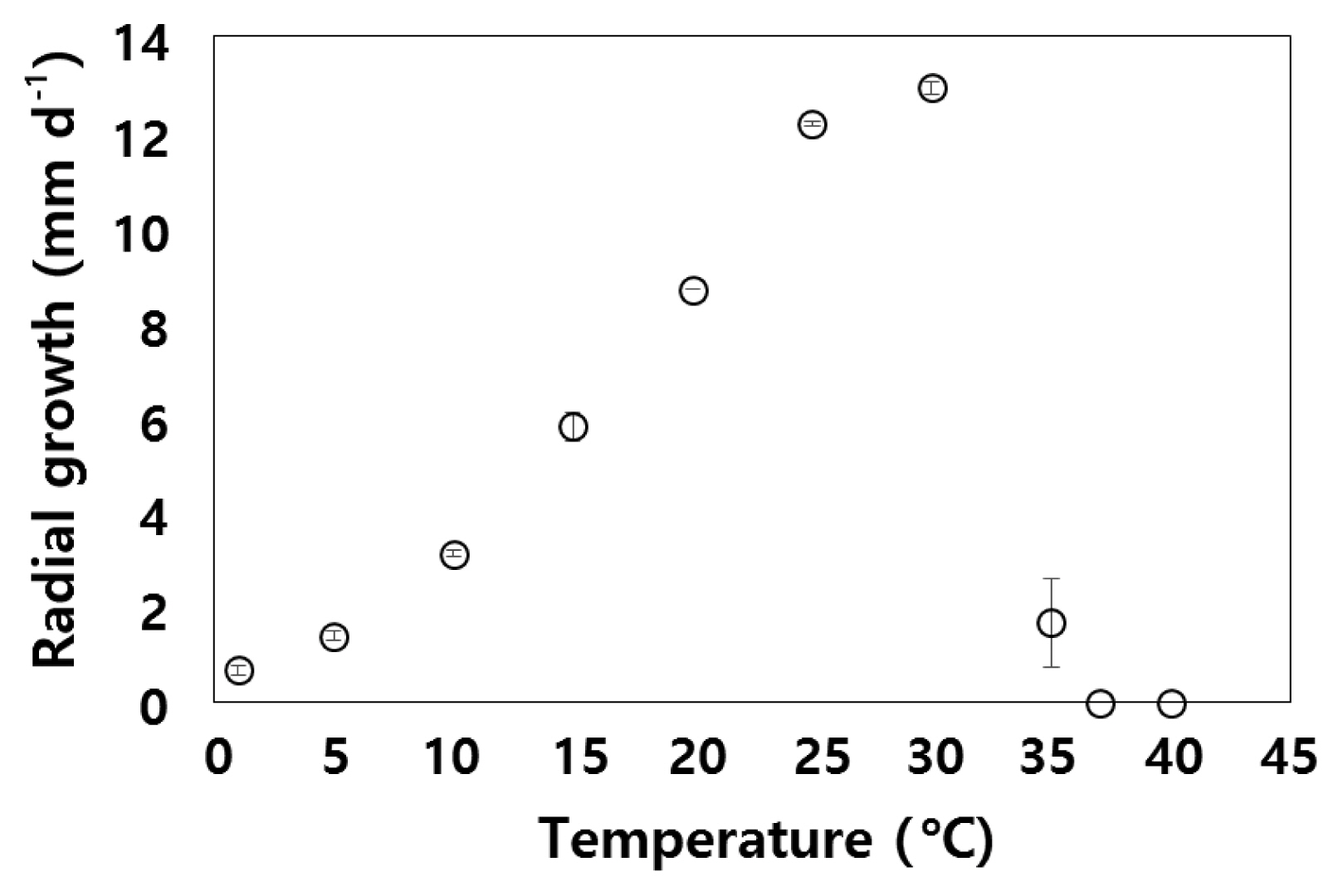
Fig. 5Effect of salinity on total radial growth of Pythium sp. isolate NIBIO 231095 after 4 days on cornmeal agar at 20°C. Values are means (± standard deviation, n = 4), ANOVA showed significant difference between salinities (p < 0.001). Letters show result of Tukey’s HSD (p = 0.05); different letters refer to significant differences between means. 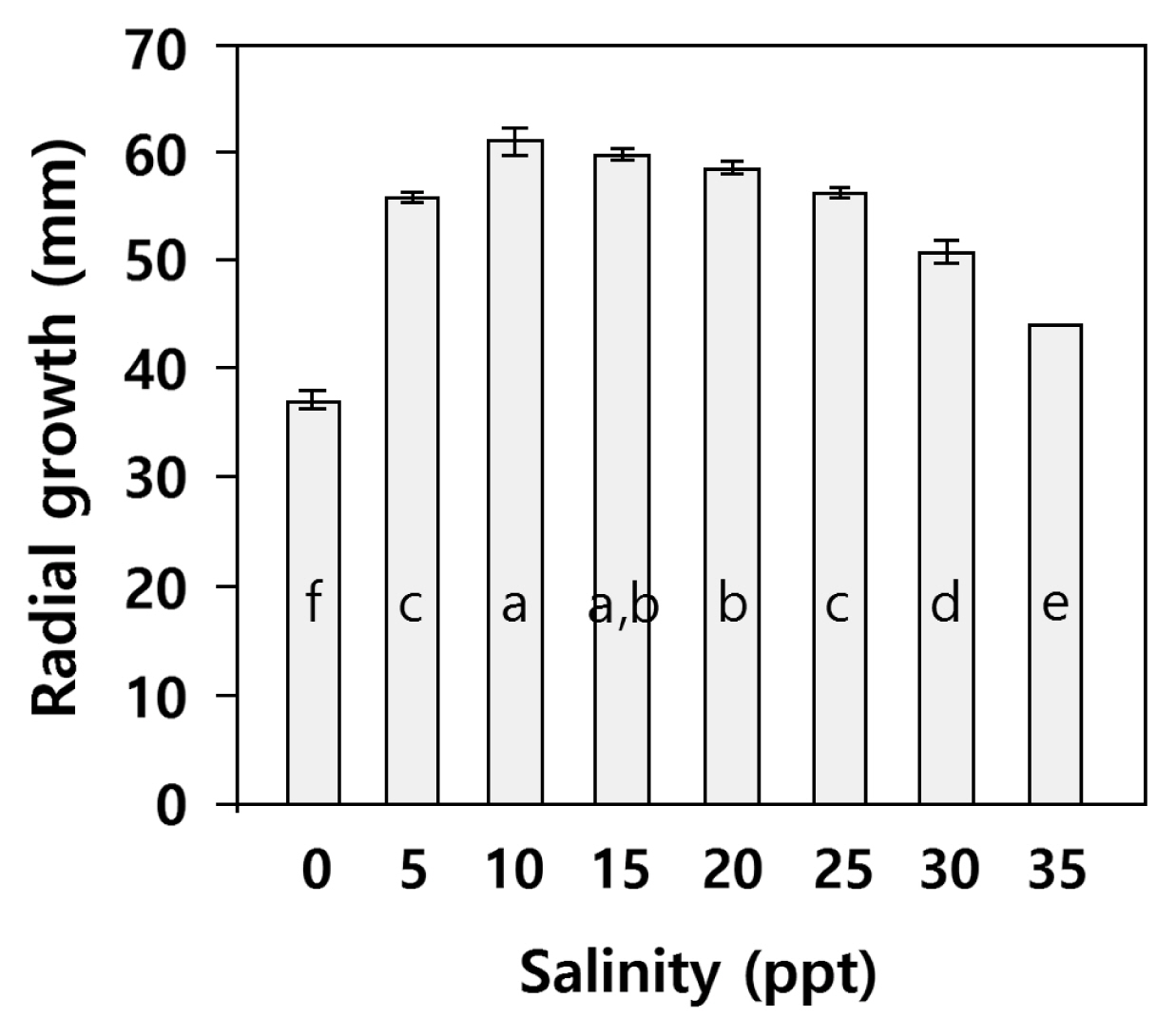
REFERENCESAleem, AA. 1980.
Pythium marinum Sparrow (Phycomycetes) infesting Porphyra leucosticta Thuret in the Mediterranean Sea. Bot Mar. 23:405–407.
Arasaki, S. 1947. Studies on the rot of Porphyra tenera by Pythium
. Nippon Suisan Gakkaishi. 13:74–90, (in Japanese)..
Bala, K., Robideau, GP., Lévesque, CA., De Cock, AWAM., Abad, ZG., Lodhi, AM., Shahzad, S., Ghaffar, A. & Coffey, MD. 2010.
Phytopythium Abad, de Cock, Bala Robideau, Lodhi & Lévesque, gen. nov. and Phytopythium Sindhum Lodhi, Shahzad and Levesque, sp. nov. Persoonia. 24:136–137.
Cavalier-Smith, T. 2018. Kingdom Chromista and its eight phyla: a new synthesis emphasising periplastid protein targeting, cytoskeletal and periplastid evolution, and ancient divergences. Protoplasma. 255:297–357.
Czeczuga, B. 1996. Species of Pythium isolated from eggs of fresh-water fish. Acta Mycol. 31:151–161.
Davis, KS. 2016. Biodiversity of aquatic oomycetes in the Falkland Islands. PhD dissertation. University of Aberdeen, Aberdeen, 266 pp.
De Cock, AWAM. 1986. Marine Pythiaceae from decaying seaweeds in the Netherlands. Mycotaxon. 25:101–110.
De Cock, AWAM. & Lévesque, CA. 2004. New species of Pythium and Phytophthora.
. Stud Mycol. 50:481–487.
Diehl, N., Kim, GH. & Zuccarello, GC. 2017. A pathogen of New Zealand Pyropia plicata (Bangiales, Rhodophyta), Pythium porphyrae (Oomycota). Algae. 32:29–39.
Dumilag, RV. 2019. Detection of Pythium porphyrae infecting Philippine Pyropia acanthophora based on morphology and nuclear rRNA internal transcribed spacer sequences. J Gen Plant Pathol. 85:72–78.
Edwards, DM., Reed, RH. & Stewart, WD. 1988. Osmoacclimation in Enteromorpha intestinalis: long-term effects of osmotic stress on organic solute accumulation. Mar Biol. 98:467–476.
Fell, JW. & Master, IM. 1975. Phycomycetes (Phytophthora spp. nov. and Pythium sp. nov.) associated with degrading mangrove (Rhizophora mangle) leaves. Can J Bot. 53:2908–2922.
Ferguson, AJ. & Jeffers, SN. 1999. Detecting multiple species of Phytophthora in container mixes from ornamental crop nurseries. Plant Dis. 83:1129–1136.
Gaastra, W., Lipman, LJ., De Cock, AW., Exel, TK., Pegge, RB., Scheurwater, J., Vilela, R. & Mendoza, L. 2010.
Pythium insidiosum: an overview. Vet Microbiol. 146:1–16.
Guiry, MD. & Guiry, GM. 2019. AlgaeBase, World-wide electronic publication, National University of Ireland, Galway, Available from: http://www.algaebase.org
. Accessed May 15, 2019
Hayden, HS., Blomster, J., Maggs, CA., Silva, PC., Stanhope, MJ. & Waaland, JR. 2003. Linnaeus was right all along: Ulva and Enteromorpha are not distinct genera. Eur J Phycol. 38:277–294.
Kamer, K. & Fong, P. 2001. Nitrogen enrichment ameliorates the negative effects of reduced salinity on the green macroalga Enteromorpha intestinalis
. Mar Ecol Prog Ser. 218:87–93.
Kang, EJ., Kim, J-H., Kim, K., Choi, H-G. & Kim, KY. 2014. Re-evaluation of green tide-forming species in the Yellow Sea. Algae. 29:267–277.
Kazama, FY. & Fuller, MS. 1977. Colonization of Porphyra perforata thallus discs by Pythium marinum, a marine facultative parasite. Mycologia. 69:246–254.
Kerwin, JL., Johnson, LM., Whisler, HC. & Tuininga, AR. 1992. Infection and morphogenesis of Pythium marinum in species of Porphyra and other red algae. Can J Bot. 70:1017–1024.
Kim, GH., Moon, K-H., Kim, J-Y., Shim, J. & Klochkova, TA. 2014. A revaluation of algal diseases in Korean Pyropia (Porphyra) sea farms and their economic impact. Algae. 29:249–265.
Klochkova, TA., Jung, S. & Kim, GH. 2017. Host range and salinity tolerance of Pythium porphyrae may indicate its terrestrial origin. J Appl Phycol. 29:371–379.
Kurokawa, K. & Tojo, M. 2010. First record of Pythium grandisporangium in Japan. Mycoscience. 51:321–324.
Kuzmina, ML., Johnson, KL., Barron, HR. & Hebert, PDN. 2012. Identification of the vascular plants of Churchill Manitoba, using a DNA barcode library. BMC Ecol. 12:25 pp.
Larsson, A. 2014. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics. 30:3276–3278.
Lee, SJ., Hwang, MS., Park, MA., Baek, JM., Ha, D-S., Lee, JE. & Lee, S-R. 2015. Molecular identification of the algal pathogen Pythium chondricola (Oomycetes) from Pyropia yezoensis (Rhodophyta) using ITS and cox1 markers. Algae. 30:217–222.
Lee, SJ., Jee, BY., Son, M-H. & Lee, S-R. 2017. Infection and cox2 sequence of Pythium chondricola (Oomycetes) causing red rot disease in Pyropia yezoensis (Rhodophyta) in Korea. Algae. 32:155–160.
Lévesque, CA. & De Cock, AWAM. 2004. Molecular phylogeny and taxonomy of the genus Pythium. Mycol. Res. 108:1363–1383.
Martins, I., Oliveira, JM., Flindt, MR. & Marques, JC. 1999. The effect of salinity on the growth rate of the macroalgae Enteromorpha intestinalis (Chlorophyta) in the Mondego estuary (west Portugal). Acta Oecol. 20:259–265.
McAvoy, KM. & Klug, JL. 2005. Positive and negative effects of riverine input on the estuarine green alga Ulva intestinalis (syn. Enteromorpha intestinalis) (Linneaus). Hydrobiologia. 545:1–9.
Park, CS., Kakinuma, M. & Amano, H. 2001. Detection of the red rot disease fungi Pythium spp. by polymerase chain reaction. Fish Sci. 67:197–199.
Park, CS., Sakaguchi, K., Kakinuma, M. & Amano, H. 2000. Comparison of the morphological and physiological features of the red rot disease fungus Pythium sp. isolated from Porphyra yezoensis from Korea and Japan. Fish Sci. 66:261–269.
Rambaut, A. & Drummond, A. 2016. Figtree version 1.4. 3, Institute of Evolutionary Biology, University of Edinburgh, Edinburgh, Available from: http://tree.bio.ed.ac.uk/software/figtree
. Accessed May 15, 2019
Reed, RH. & Russell, G. 1979. Adaptation to salinity stress in populations of Enteromorpha intestinalis (L.) Link. Estuar Coast Mar Sci. 8:251–258.
Ritchie, RJ. & Larkum, AWD. 1985. Potassium transport in Enteromorpha intestinalis (L) Link: II Effects of medium composition and metabolic inhibitors. J Exp Bot. 36:394–412.
Robideau, GP., De Cock, AWAM., Coffey, MD., Voglmayr, H., Brouwer, H., Bala, K., Chitty, DW., Désaulniers, N., Eggertson, QA., Gachon, CMM., Hu, C., Küpper, FC., Rintoul, TL., Sarhan, E., Verstappen, ECP., Zhang, Y., Bonants, PJM., Ristaino, JB. & Lévesque, CA. 2011. DNA barcoding of oomycetes with cytochrome c oxidase subunit I and internal transcribed spacer. Mol Ecol Resour. 11:1002–1011.
Shimada, S., Yokoyama, N., Arai, S. & Hiraoka, M. 2008. Phylogeography of the genus Ulva (Ulvophyceae, Chlorophyta), with special reference to the Japanese freshwater and brackish taxa. J Appl Phycol. 20:979–989.
Sparrow, FK. Jr 1934. Observations on marine Phycomycetes collected in Denmark. Dansk Bot Ark. 8:1–24.
Stamatakis, A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
Takahashi, M., Ichitani, T. & Sasaki, M. 1977.
Pythium porphyrae Takahashi et Sasaki, sp. nov. causing red rot of marine red algae Porphyra spp. Trans Mycol Soc Jpn. 18:279–285.
Thompson, TA. 1982. Some aspects on the taxonomy, ecology and histology of Pythium Pringsheim species associated with Fucus distichus in estuaries and marine habitats of British Columbia. PhD dissertation. University of British Columbia, Vancouver, BC, 114 pp.
Uppalapati, SR. & Fujita, Y. 2000. Carbohydrate regulation of attachment, encystment, and appressorium formation by Pythium porphyrae (Oomycota) zoospores on Porphyra yezoensis (Rhodophyta). J Phycol. 36:359–366.
Uzuhashi, S., Kakishima, M. & Tojo, M. 2010. Phylogeny of the genus Pythium and description of new genera. Mycoscience. 51:337–365.
Uzuhashi, S., Okada, G. & Ohkuma, M. 2015. Four new Pythium species from aquatic environments in Japan. Antonie Van Leeuwenhoek. 107:375–391.
Van der Plaats-Niterink, AJ. 1981. Monograph of the genus Pythium
. Stud Mycol. 21:1–242.
|
|
||||||||||||||||||||||||||||||||||||||||||