ABSTRACTThis study provides the first report of the presence of Coolia malayensis in the Mediterranean Sea, co-occurring with C. monotis. Isolated strains from the Gulf of Gabès, Tunisia (South-eastern Mediterranean) were identified by morphological characterization and phylogenetic analysis. Examination by light and scanning electron microscopy revealed no significant morphological differences between the Tunisian isolates and other geographically distant strains of C. monotis and C. malayensis. Phylogenetic trees based on ITS1-5.8S-ITS2 and D1–D3/28S rDNA sequences showed that C. monotis strains clustered with others from the Mediterranean and Atlantic whereas the C. malayensis isolate branched with isolates from the Pacific and the Atlantic, therefore revealing no geographical trend among C. monotis and C. malayensis populations. Ultrastructural analyses by transmission electron microscopy revealed the presence of numerous vesicles containing spirally coiled fibers in both C. malayensis and C. monotis cells, which we speculate to be involved in mucus production.
INTRODUCTIONThe genus Coolia belongs to the family of Ostreopsidaceae (Gonyaucales, Dinophyceae), which includes two genera Ostreopsis and Coolia. The type species C. monotis Meunier was first described in Nieuport (North Sea, Belgium) (Meunier 1919) and it was many decades after the generic type description that C. tropicalis Faust (Faust 1995) was added to the genus. In the following years, six other species have been described: C. areolata Ten-Hage, Turquet, Quod & Couté (Ten-Hage et al. 2000), C. canariensis S. Fraga (Fraga et al. 2008), C. malayensis Leaw, P-T. Lim & Usup (Leaw et al. 2010), C. santacroce, C. palmyrensis Karafas, Tomas & R. York (Karafas et al. 2015), and C. guanchica sp. nov. H. David, Laza-Martínez, F. Rodríguez & S. Fraga (David et al. 2019).
The taxonomy identification of Coolia to species level is mainly based on the morphological characters, such as thecal plate pattern, shape and size revealed by both light and scanning electron microscopy (Ten-Hage et al. 2000, Fraga et al. 2008, Leaw et al. 2010, Karafas et al. 2015, David et al. 2019). However, the presence of morphologically cryptic species, as is the case of C. monotis species complex (Leaw et al. 2010, 2016, Momigliano et al. 2013, Karafas et al. 2015) creates misidentification. Therefore, a molecular phylogenetic approach is often used to complement morphology. At present, molecular data is available for all Coolia species (Penna et al. 2005, Fraga et al. 2008, Leaw et al. 2010, Momigliano et al. 2013, Karafas et al. 2015) except for C. areolata. Different ribosomal DNA (rDNA) gene markers have been used to support species delineation namely the large subunit (LSU) (Dolapsakis et al. 2006, Fraga et al. 2008, Leaw et al. 2010, Karafas et al. 2015, Ben-Gharbia et al. 2016, Gómez et al. 2016, David et al. 2019, Tibiriçá et al. 2020, Zhang et al. 2020), the 5.8S and the internal transcribed spacers (ITS) (Penna et al. 2005, Leaw et al. 2010, David et al. 2014, 2019, Karafas et al. 2015, Nascimento et al. 2019, Tibiriçá et al. 2020, Zhang et al. 2020) and the small subunit (Momigliano et al. 2013, Wakeman et al. 2015). The ITS2 secondary structure has also been documented to be useful in differentiating between Coolia species (Leaw et al. 2010, 2016).
Species of Coolia can live in diverse environments and are globally distributed from temperate to tropical waters. All Coolia species except C. monotis, C. areolata, and C. santacroce have been observed in the Pacific Ocean, particularly in the west (Faust 1995, Holmes et al. 1995, Fraga et al. 2008, Leaw et al. 2010, Mohammad-Noor et al. 2013, Momigliano et al. 2013, Rhodes et al. 2014b, Karafas et al. 2015, Larsson et al. 2019, Zhang et al. 2020). C. canariensis and C. monotis have been reported in the eastern and western Atlantic (Penna et al. 2005, Fraga et al. 2008, Laza-Martínez et al. 2011, David et al. 2014, Leaw et al. 2016, Lewis et al. 2018, Mendes et al. 2019, Nascimento et al. 2019), while records of C. malayensis, C. palmyrensis, C. santacroce, and C. tropicalis have been restricted to the western Atlantic (Faust 1995, Karafas et al. 2015, Leaw et al. 2016, Mendes et al. 2019, Nascimento et al. 2019, Tibiriçá et al. 2020) and C. guanchica to the eastern Atlantic (David et al. 2019). C. areolata has only been detected in the Southwest Indian Ocean (Ten-Hage et al. 2000). In the Mediterranean Sea, C. monotis is widely dispersed from the western (Halim 1960, Vila et al. 2001, Penna et al. 2005) to the eastern basin (Aligizaki and Nikolaidis 2006, Armi et al. 2010, Ismael 2014, Ben-Gharbia et al. 2016, Abdennadher et al. 2020).
Great attention has been given to the genus Coolia since an Australian isolate named C. monotis was associated with cooliatoxin, a yessotoxin analog that induced hypothermia and respiratory failure in mice (Holmes et al. 1995). Later it was re-identified as C. malayensis (Mohammad-Noor et al. 2013, Rhodes et al. 2014b). C. malayensis, C. tropicalis, C. palmyrensis, and C. santacroce have been reported to be toxic and biotoxin production was confirmed using cytotoxicity bioassays, hemolytic assays and chemical analysis (Holmes et al. 1995, Rhodes and Thomas 1997, Rhodes et al. 2014a, Karafas et al. 2015, Wakeman et al. 2015, Leung et al. 2017, Tibiriçá et al. 2020). Moreover, it has been shown that C. malayensis can be source of bioactive compounds (Shah et al. 2014).
In the literature, Coolia have been isolated from various substrates, including macroalgae (Mohammad-Noor et al. 2013, Momigliono et al. 2013, Tawong et al. 2015, David et al. 2019, Tibiriçá et al. 2020, Zhang et al. 2020), floating macroalgae (Rhodes et al. 2014b), turf algae (Wakeman et al. 2015), seagrasses (Leaw et al. 2010, Rhodes et al. 2014b), dead corals (Leaw et al. 2010, Leung et al. 2017), rock surfaces (Leung et al. 2017), sand (Faust 1995, Leaw et al. 2010), tide pools (Faust 1995), and plastic screens (Faust 1995, Karafas et al. 2015). However, only Mohammad Noor et al. (2019) studied the substrate preference of Coolia species and reported their preference to brown macroalgae Sargassum and Padina.
In our study area, the Gulf of Gabès (Fig. 1), located in the South-eastern Mediterranean Sea (Southeast of Tunisia) and sheltering Kerkennah and Djerba islands and Boughrara and El Bibane lagoons, C. monotis has been recorded in the water column (Abdennadher et al. 2020), attached to the sediment and the adjacent water “biofilm” (Loukil-Baklouti et al. 2018) and on various substrates (magnoliophytes and macroalgae) (Moncer et al. 2017).
In the Gulf of Gabès, little is known about the diversity of Coolia species. The primary aim of this study was to isolate, identify and characterize the Coolia species from this area. Fifteen strains of Coolia spp. were established into clonal cultures and their taxonomic identity determined through morphological and phylogenetic examination. Morphology was examined by light microscopy (LM), scanning electron microscopy (SEM), and transmission electron microscopy (TEM), and the ITS region containing the 5.8S rDNA and the D1–D3 LSU rDNA were used for phylogenetic analysis.
MATERIALS AND METHODSSites description and samplingWater samples were collected from shallow sites (1 m depth) during the spring tide when the tide coefficient was the highest (>1.50 m) using a Van Dorn bottle at three geographically distinct sites along the Gulf of Gabès in southern Tunisia (South-eastern Mediterranean) (Fig. 1). The first site, Cercina (34°41′46″ N, 11°07′24″ E) is located on the western coast of Kerkennah Islands, situated north of the Gulf of Gabès. The second, Boughrara lagoon (33°39′10″ N, 10°43′58″ E) is on the North coast of Medenine city in the south of the Gabès Gulf and is a quasi-closed paralic environment, connected to the Mediterranean via two Channels, the El Kantara Channel (width: 5 km, average depth: 1 m) to the northeast and the Ajim Channel to the northwest (width: 2.2 km, average depth: 15 m). The third site, Cotusal Channel (33°22′48″ N, 11°08′24″ E), is positioned in the coastal region of Zarzis, towards the south of the Gulf of Gabès, on the Lemsa plateau which is surrounded by two salt flats (El Melah and Boujmel). All sampling sites were characterized by the presence of P. oceanica seagrass beds (Hattour and Ben Mustapha 2013) and colonized by many other macrophytes known as hosts of different epiphytic species. Photophylic algae such as Cystoseira, Padina pavonica, and Caulerpa prolifera were present at the Cercina site (Hattour and Ben Mustapha 2013) while in the Boughrara lagoon, the high densities of C. nodosa, C. prolifera, and Cystoseira were recorded (Hattour and Ben Mustapha 2013). On the Zarzis coasts, C. nodosa, C. prolifera, and C. racemosa were observed and Penicelius capitatus and Halimeda tuna are also present (Hattour and Ben Mustapha 2013).
Cell isolation
Coolia cells were isolated from collected water samples using the micropipette technique (Andersen 2005) under an inverted LM (CK40; Olympus, Tokyo, Japan) into 96-multiwell culture plates (Sigma-Aldrich, St. Louis, MO, USA) in L1 medium (Guillard and Hargraves 1993) made with seawater from the Gulf of Gabès and cultures were subsequently transferred to a Nunclon culture flasks once established (Sigma-Aldrich). Clonal cultures of fourteen C. monotis (Com.2–Com.8, Com.10–Com.16) strains, and one C. malayensis (Com.1) were established (Supplementary Table S1). The strains Com.1 to Com.11 and Com.13 to Com.16 were isolated from the Cotusal Channel and Boughrara Lagoon sites, respectively. The Com.12 strain was isolated from the Cercina site. Cultures were maintained at salinity 40 in L1 medium at 22°C, on a 12 : 12 h light : dark cycle under an irradiance of 100 μmol photons m−2 s−1.
Morphological analysesMorphometric features (cell length: the distance between apex and antapex or anteroposterior; width: the distance between the lateral sides) and thecal plates were examined under LM and SEM. The ultrastructural analyses were performed with a TEM. In this study, a tabulation system as described in Balech (1956) was followed to name the plates. LM observations were carried out on living or lugol-fixed cultured cells using a Carl Zeiss Microscopy GmbH (Jena, Germany) at ×100, ×200, and ×400 magnifications. Images were collected using an Axiocam 105 color digital camera (Carl Zeiss) with capture software (ZEN core v2.7 acquisition and analysis; Carl Zeiss). For SEM observations, samples were fixed with formaldehyde at 1% final concentration, filtered on 3 μm pore size polycarbonate filters (Nuclepore, Pleasanton, CA, USA), rinsed with distilled water, dehydrated in an ethanol series (25, 50, 75, 95, and 100%) and critical-point-dried. The filters were mounted on stubs, sputter-coated with gold-palladium and observed with a JEOL JSM-6500F SEM (JEOL-USA Inc., Peabody, MA, USA).
Cells ultrastructure were examined under a transmission electron microscope Morgagni 268D (FEI, Eindhoven, Netherlands) as described in Abdennadher et al. (2017). Digital images were acquired with a CCD Mega-View (SIS, Münster, Germany).
Molecular analysesDNA extraction, polymerase chain reaction amplification, and sequencingTotal genomic DNA was extracted according to the protocol of Scholin et al. (1994) modified by Mikulski et al. (2005). Briefly, dinoflagellates cells were harvested by centrifugation at 7,000 rpm for 10 min and resuspended in 300 μL of Milli-Q water. To lyse the cells, 30 μL of lysozyme (10 mg mL−1) was added and the suspension incubated at 37°C for 30 min. Then, 30 μL of sodium dodecyl sulfate (10%), 3 μL of 1 M EDTA (pH 8), 3 μL of 1 M Tris-HCl (pH 7.5), 42 μL of 5 M NaCl, and 30 μL of cetrimonium bromide (10%) were added in the order given with gentle mixing after each step, and the mixture was incubated at 65°C for 10 min. The lysate was extracted with phenol / chloroform / isoamyl alcohol (25 : 24 : 1, pH 8) and then with chloroform / isoamyl alcohol (24 : 1) to remove the residual phenol. The DNA was precipitated overnight at −20°C with two volumes of absolute ethanol and 1/10 volume of sodium acetate (3 M, pH 4.8), rinsed with 70% ethanol, air-dried and resuspended in 40 μL of TE (10 mM Tris-HCl, pH 7.4; 1 mM EDTA, pH 8). The DNeasy Power Water Kit (Qiagen, Hilden, Germany) was also used and the genomic DNA was extracted following the manufacturer’s protocol. The DNA concentration and quality were determined with a NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA).
The internal transcribed spacer (ITS1-5.8S-ITS2) of the rDNA was amplified by polymerase chain reaction (PCR) with ITS1F and ITS1R primers (Adachi et al. 1994, Leaw et al. 2001). The PCR reaction (50 μL) contained 20 ng of genomic DNA as a template, 5 μL of PCR buffer, 200 μM of each deoxynucleotide triphosphate, 2 mM MgCl2, 0.2 μM of each primer, 0.15 mg mL−1 of bovine serum albumin (BSA) and 2.5 units of Taq polymerase (Invitrogen, Carlsbad, CA, USA). Thermocycler conditions included an initial denaturing step at 94°C for 3 min followed by 35 cycles of denaturing at 94°C for 45 s, annealing at 50°C for 90 s and an extension at 72°C for 90 s. A 10 min elongation step at 72°C was ultimately carried out.
PCR of the D1/D3 region of the LSU of the rDNA was conducted using Taq DNA Polymerase 2X-preMix (GeneON, Ludwigshafen, Germany) under the following program: 1 cycle of 3 min at 95°C, 35 cycles of 30 s at 95°C, 45 s at 58°C and 60 s at 72°C and 1 cycle of 6 min at 72°C. The primers D1R and D3ca (Scholin et al. 1994) were used at a final concentration of 0.35 μM. The genomic DNA and BSA were used as described above. Amplification products were further purified using the QIAquick PCR purification kit (Qiagen) according to the manufacturer’s instructions, quantified with the NanoDrop 2000 spectrophotometer and sequenced by the Eurofins Genomics DNA sequencing services (Ebserberg, Germany). Sequencing reactions were performed with the same primer pairs used for PCR amplification using the ABI BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) and were run on the DNA Engine Tetrad 2 Peltier Thermal Cycler (Bio-Rad, Hercules, CA, USA). Thirteen sequences of LSU rDNA D1/D3 and three sequences of ITS-5.8S rDNA were obtained and were deposited in GenBank under the accession numbers listed in Supplementary Table S2. Finally, the DNA sequence homology search within the GenBank database was performed using BLAST.
Alignments and phylogenetic analysesThe alignments of ITS-5.8S rDNA and LSU rDNA D1/D3 sequences of C. malayensis, C. monotis, C. tropicalis, C. canariensis, C. santacroce, C. palmyrensis, and C. guanchica (Supplementary Table S2), including the newly obtained sequences of Coolia species isolated from the Gulf of Gabès, were performed separately using MAFFT v7.471 (Madeira et al. 2019). The multiple alignments of the ITS and LSU involved 58 and 60 nucleotide sequences, respectively. The sequences of O. ovata (Oso.3) and O. siamensis (CSIC-D) were used as outgroups for the LSU rDNA D1/D3 phylogeny, while for ITS-5.8S rDNA phylogeny, two strains of O. ovata (OVPD7 and Oso.2) were chosen as outgroups. The alignments were subsequently refined by hand with the software TextPad version 8.5.1 and the final alignments of ITS-5.8S rDNA and LSU rDNA D1/D3 sequences consisted of 826 (100 conserved sites and 726 variables sites) and 520 (135 conserved sites and 385 variables sites) nucleotides, correspondingly. Phylogenetic and molecular evolutionary analyses of both rDNA regions were conducted under the software MEGA X (Kumar et al. 2018). The Tamura-Nei (TN93 + G, G = 1.351) (Tamura and Nei 1993) and the Tamura 3-parameter (T92 + G, G = 0.759) (Tamura 1992) models were selected as the best-fit models of nucleotide substitution rate to build the maximum likelihood (ML) (Felsenstein 1981) phylogenetic trees of the ITS-5.8S rDNA and the LSU rDNA D1/D3 regions, respectively. The robustness of the ML distance trees was evaluated using bootstrapping with 5,000 replications and all positions containing gaps and missing data were eliminated. The trees were visualized in MEGA X and edited using the software CorelDRAW Graphics Suite 2021 (ver. 23; Corel Corporation, Ottawa, Canada). Pairwise distances between strains and mean pairwise distances between clades were computed using MEGA X and were in the units of the number of base substitutions per site. A discrete Gamma distribution was used to model evolutionary rate differences among sites and a bootstrap procedure (5,000 replicates) was selected.
RESULTSSpecies descriptionCell morphology was described using one isolate of C. malayensis (Com.1) and fourteen isolates of C. monotis (Com.2–Com.8, Com.10–Com.16). The cells of C. malayensis strain Com.1 were spherical in dorsoventral view and ellipsoid in lateral view, with a smooth surface covered by scattered pores (Figs 2A, B & 3). Cells measured 22 to 26 μm in length (23.3 ± 2.1 μm, n = 50) and 25 to 30.9 μm in width (30.6 ± 2.0 μm, n = 50) (Table 1). The plate tabulation follows the same formula as for other Coolia species: Po, 3′, 7″, 6C, ?S, 5‴, 2‴′ (Leaw et al. 2010). The apical pore plate (Po) was slightly curved with a length varying between 4 and 7.3 μm (5.3 ± 0.9 μm, n = 10) (Fig. 3A & D). Plate 1′ was elongated and hexagonal (Fig. 3B). The 3′ plate was quadrangular, bordering the 1′, 2′, 4″ and 6″ and did not touch the 5″ (Fig. 3A). The 6″ plate was the biggest epithecal plate, pentagonal and occupied nearly half of the epitheca (Fig. 3A & B). The seventh precingular plate 7″ was pentagonal with a width-length ratio ranging from 1.2 to 1.5 μm (1.3 ± 1.3 μm, n = 10) (Fig. 3B & E). The 1‴ plate was triangular and was the smallest plate of the hypotheca. The 3‴ was the largest plate in the hypotheca and touched the 2‴, 4‴, 1‴′, and 2‴′ (Fig. 3C & D). The 2‴′ was quadrangular, touching the sulcal plate and was separated of the first 1‴′ antapical plate by the sulcus (Fig. 3D).
The microscopic study showed that C. monotis cells were round, lens-shaped and anteroposteriorly compressed (Figs 2C–F & 4). The size of cells ranged from 24.6 to 39.7 μm in length (31.36 ± 3.56 μm, n = 700) and from 26 to 40.9 μm in width (33.65 ± 3.49 μm, n = 700) (Table 1). The thecal surface was covered with well-defined plates (Fig. 4). The plate formula of C. monotis was Po, 3′, 7″, 6C, ?S, 5‴, and 2‴′. On the epitheca, which was slightly smaller than the hypotheca, a distinct Po was positioned off-center and was located adjacent to the apical plates 1′, 2′, and 3′ (Fig. 4B & C). The Po was slightly curved with a length ranging from 5 to 9.2 μm. Plate 1′ was oblong, touching plate 6″ and hexagonal with its right side located in the middle of the dorsoventral part (Fig. 4C). Plate 2′ was hexagonal, elongated and overlapped by plates 3′, 2″, 3″, and 4″ (Fig. 4C). The 3′ plate was pentagonal, situated centrally in the dorsal part of the epitheca and touches Po, 1′, 2′, 4″, 5″, and 6″ (Fig. 4B). Plate 2″ was wider than its neighboring precingular plates 1″ and 3″ (Fig. 4C). Off the precingular plate series, 6″ was the largest, pentagonal and it occupied nearly half of the epitheca (Fig. 4D). Plate 7″ was pentagonal (Fig. 4E) with a width / length ratios ranging from 1 to 1.7. The hypotheca was composed of five postcingular plates (5‴) and two antapical plates (2‴′) (Fig. 4F). Plate 1‴ was the smaller postcingular plate (Fig. 4F). Plates 2‴ and 4‴ were equal in size (Fig. 4F). Plate 3‴ was quadrangular and it was equal or larger than the 4‴ plate (Fig. 4F). The 1‴ and 5‴ plates were triangular (Fig. 4E). Plate 2‴′ was pentagonal and smaller than the 1‴′ (Fig. 4F).
TEM micrographs showed that both species presented the typical dinoflagellate ultrastructure including a nucleus (Fig. 5A & B) surrounded by a regular nuclear membrane with nuclear pores (Fig. 5B), the permanently condensed chromosomes (Fig. 5B), numerous chloroplasts containing 1 to 2 pyrenoids (Fig. 5B), trichocysts (T) (Fig. 5C & E), lipid bodies, starch grains, and golgi bodies (Fig. 5C & F). It was also observed the presence of many spirally coiled fibers (SCFs) containing vesicles (VE) throughout the cytoplasm (Fig. 5E). The amphiesma was composed of flattened amphiesmal VE within plate-like material (Fig. 5D).
Phylogenetic analysesPhylogenetic trees inferred from LSU rDNA D1/D3 (Fig. 6) and ITS-5.8S rDNA (Fig. 7) regions delineated seven distinct clades corresponding to named species, C. monotis, C. malayensis, C. santacroce, C. palmyrensis, C. tropicalis, C. canariensis, and C. guanchica. Coolia strains collected from the Gulf of Gabès, Tunisia (South-eastern Mediterranean) branched into two distinct clades, the strain C. malayensis Com.1 clustered with a well-supported bootstrap (99%) with sequences of this species from Pacific and Atlantic oceans (Fig. 6) and C. monotis isolates (Com.2–Com.4, Com.6–Com.8, and Com.11–Com.16) were grouped with high bootstrap with those from Mediterranean Sea (Greece, Italian, and Spanish coasts) and Atlantic Ocean (Rhode Island and Western European coasts) (Figs 6 & 7). C. santacroce, C. palmyrensis, C. tropicalis, C. canariensis, and C. guanchica clades were well distinguished and were supported by high bootstraps (Figs 6 & 7). C. monotis, C. malayensis, C. santacroce, and C. palmyrensis clustered together as sister taxa with 99% bootstrap support for both LSU and ITS rDNA phylogenies. Indeed, the lowest interspecific evolutionary divergence was detected between C. monotis and C. santacroce for LSU (0.122 ± 0.015) and between C. monotis and C. malayensis for ITS (0.181 ± 0.015) (Tables 2 & 3). C. palmyrensis is closely related to C. santacroce for LSU (0.156 ± 0.016) and to C. monotis for ITS (0.242 ± 0.017) (Tables 2 & 3). The average evolutionary divergences of the aligned ITS-5.8S rDNA and LSU rDNA D1/D3 sequences were 0.24 ± 0.01 and 0.17 ± 0.01, respectively.
The highest genetic distance was detected within C. canariensis clade for both ITS (0.31 ± 0.01) and LSU (0.12 ± 0.01) alignments (Tables 2 & 3). The Tunisian strain C. malayensis Com.1 exhibited the lower p-distance values with C. malayensis strains CmPD22 (0.0055) from Port Dickson, Malaysia and CAWD154 (0.014) from New Zealand for LSU and ITS regions, respectively (Leaw et al. 2016).
DISCUSSIONSpecies descriptionUp until now, the presence of C. malayensis has only been reported in the North and south parts of the Atlantic and Pacific Oceans and in the Caribbean Sea (Leaw et al. 2010, Rhodes et al. 2014b, Karafas et al. 2015, Tawong et al. 2015, Wakeman et al. 2015, Gómez et al. 2016, Mendes et al. 2019, Nascimento et al. 2019, Tibiriçá et al. 2020, Zhang et al. 2020). To our knowledge, this study presents the first record of C. malayensis in the Mediterranean Sea, also providing its detailed description. Cell measurements (Table 4) and the apical pore length (4–7.3 μm) of the Com.1 strain were similar to those found in other studies (Leaw et al. 2010, 2016, Karafas et al. 2015, Tawong et al. 2015, Gómez et al. 2016, Mendes et al. 2019, Nascimento et al. 2019, Tibiriçá et al. 2020, Zhang et al. 2020). For the local strain from the South-east Mediterranean Sea, the 3‴ plate is larger than the 4‴ plate, which fits well with that described for C. malayensis strains from Pacific and West Atlantic Oceans (Table 4). Similarly, to the Malaysian (Leaw et al. 2010, 2016), Puerto Rico and Brazilian (Gómez et al. 2016) isolates, the shape of the 3′ plate was quadrangular, while previous works observed a pentagonal 3′ plate (Table 4). Yet, Karafas et al. (2015) noted pentagonal or hexagonal 3′ plate forms. Moreover, the width / length ratio of 7″ plate (1.2–1.5) was in the range of the reported strains (Table 4).
The size of cells from the C. monotis strain isolated from the Gulf of Gabès (South-eastern Mediterranean) was comparable to that of C. monotis cells previously described from all over the world (Meunier 1919, Penna et al. 2005, Dolapsakis et al. 2006, Laza-Martínez et al. 2011, Pagliara and Caroppo 2012, David et al. 2014, Ho and Nguyen 2014, Karafas et al. 2015, Ben-Gharbia et al. 2016, Lewis et al. 2018) (Table 4). Though, the largest C. monotis cells have been described from field samples from the Caribbean Sea (Faust 2009), the Gulf of Tunis (Armi et al. 2010) and the Arab Gulf (Al-Yamani and Saburova 2010) (Table 4). The apical pore length of the Tunisian C. monotis strains (5–9.2 μm) was within the range of that for C. monotis strains isolated from Greece (6 μm) (Aligizaki and Nikolaidis 2006), Spain (7–9 μm) (Laza-Martínez et al. 2011), Iberian Peninsula (6–9 μm) (David et al. 2014), Rhode Island (5.7–8.8 μm) (Karafas et al. 2015), and Canada (6.0–9.3 μm) (Lewis et al. 2018); but, smaller than those reported from Northwestern Mediterranean Sea (9–11 μm) (Penna et al. 2005), North Aegean Sea (10–12 μm) (Dolapsakis et al. 2006), Gulf of Tunis (9–11 μm) (Armi et al. 2010) and Northern Ionian Sea (9–11 μm) (Pagliara and Caroppo 2012) (Table 4).
The thecal plate pattern of C. monotis strains was in line with previous descriptions (Balech 1956, Penna et al. 2005, Dolapsakis et al. 2006, Laza-Martínez et al. 2011, Pagliara and Caroppo 2012, Ho and Nguyen 2014, Karafas et al. 2015, Lewis et al. 2018), but differed from the original (Meunier 1919). Indeed, Meunier (1919) showed that plates 6″ and 5″ were elongated and the suture between them running in a nearly dorsoventral direction, while for the strains isolated from the Gulf of Gabès, it runs to the right side resulting in wide 6″ and small 5″ plates. Similarly, C. monotis strains isolated from Greece (Dolapsakis et al. 2006) and Spain (Laza-Martínez et al. 2011), show the plate 2″ wider than 1″; but, in the original description, these plates exhibited a similar size (Meunier 1919). Our observations revealed a narrow 1′ plate, which was consistent with former descriptions (Dolapsakis et al. 2006, Ho and Nguyen 2014, Karafas et al. 2015). Though, Penna et al. (2005) and Pagliara and Caroppo (2012) observed a wide 1′ plate. Furthermore, Laza-Martínez et al. (2011) found that the 1′ plate of C. monotis cells from Biscay Bay, was either wide or narrow. The range of width / length ratio of plate 7″ and the pentagonal shape of the 3′ plate of C. monotis strains described in the present study corresponded to those reported by earlier studies (Penna et al. 2005, Dolapsakis et al. 2006, David et al. 2014, Karafas et al. 2015, Lewis et al. 2018) (Table 4). As already described (Karafas et al. 2015), our observations revealed that the plate 3‴ was either equal (Aligizaki and Nikolaidis 2006, Al-Yamani and Saburova 2010, David et al. 2014, Leaw et al. 2016, Lewis et al. 2018) or larger (Meunier 1919, Balech 1956, Penna et al. 2005, Dolapsakis et al. 2006, Laza-Martínez et al. 2011, Pagliara and Caroppo 2012) than the 4‴ plate.
The ultrastructural analysis of C. monotis and C. malayensis cells revealed the typical dinoflagellate cellular organization, as well as the presence of vesicles (VE) enclosing spirally coiled fibrous material (SCFs) in the cytoplasm of both species. Besada et al. (1982) recognized for the first time that benthic dinoflagellates, such as Gambierdiscus, Ostreopsis, and Coolia show many typical dinoflagellate features, but reveal the presence of a previously undescribed organelle consisting of an array of VE containing fibrous material; they suggested a link between this organelle and the enormous amount of mucilage secreted. Afterwards, it was shown that the Ostreopsis mucilage shows a complex structure, formed by a network of long fibers, derived from trichocysts extruded through thecal pores and by an amorphous matrix of acidic polysaccharides (Honsell et al. 2013). Mucilage increases during cell proliferation, producing a typical brownish mat, visible with the naked eye (Lewis et al. 2018). In fact, C. monotis Com.10 strain growing in exponential phase produced large quantities of mucilage, forming aggregates of cells that were visible in the culture (Fig. 2E & F) (Abdennadher et al. 2020). Studies focusing on the mucilaginous matrix suggest its key-role in growth strategy, defense against grazing, increased buoyancy, metabolic self-regulation (Reynolds 2007) and active role in conveying toxicity (Giussani et al. 2015). However, further Coolia strains should be examined to better understand the structure and function(s) of these SCFs.
Phylogenetic analysis of Coolia speciesThe described phylogenetic resolution discriminating the species C. monotis, C. malayensis, C. santacroce, C. palmyrensis, C. tropicalis, C. canariensis, and C. guanchica was consistent with previous studies based on LSU (Rhodes et al. 2014b, Karafas et al. 2015, Leaw et al. 2016, Leung et al. 2017, Lewis et al. 2018, David et al. 2019, Larsson et al. 2019, Mendes et al. 2019, Nascimento et al. 2019, Tibiriçá et al. 2020, Zhang et al. 2020) and ITS rDNA regions (Karafas et al. 2015, Leaw et al. 2016, David et al. 2019, Nascimento et al. 2019, Tibiriçá et al. 2020, Zhang et al. 2020). LSU and ITS phylogenetic analyses conducted in this study yielded similar clustering and showed that Coolia strains collected from the Gulf of Gabès were grouped into two clades related to C. monotis and C. malayensis species. The grouping of C. monotis strains with Mediterranean and Northeast Atlantic strains was in accordance with previous molecular phylogenetic analyses using ITS and / or LSU rDNA regions (Penna et al. 2005, Fraga et al. 2008, David et al. 2014, 2019, Karafas et al. 2015, Gómez et al. 2016, Leaw et al. 2016, Leung et al. 2017, Lewis et al. 2018, Larsson et al. 2019, Mendes et al. 2019, Nascimento et al. 2019, Tibiriçá et al. 2020, Zhang et al. 2020) and supported the presence of a single evolutionary lineage of Mediterranean and Atlantic C. monotis isolates (Penna et al. 2005), in one hand, and the North Atlantic origin of C. monotis, previously postulated by Leaw et al. (2016), in the other hand. The LSU rDNA D1/D3 sequencing and phylogeny confirmed the identity of the Tunisian isolate Com.1 as C. malayensis, previously identified as Coolia sp. This finding is consistent with an earlier study revealing the clustering of Com.1 strain in the C. malayensis clade of the ITS2 tree (Leaw et al. 2016) and demonstrated, for the first time, the occurrence in the South-eastern Mediterranean waters of C. malayensis species, previously described in the Indo-Pacific region and Brazilian waters. The clustering of the South-eastern Mediterranean strain Com.1 with C. malayensis isolates from very distant localities namely the Pacific and the Atlantic revealed no geographical genetic differentiation among C. malayensis clade, which is in accordance with many investigations using LSU and ITS rDNA genes, and ITS-2 sequence structure (Gómez et al. 2016, Leaw et al. 2016, Leung et al. 2017, Lewis et al. 2018, Larsson et al. 2019, Mendes et al. 2019, Nascimento et al. 2019, Tibiriçá et al. 2020, Zhang et al. 2020). Phylogenetic results support the C. monotis complex (C. monotis, C. malayensis, C. palmyrensis, and C. santacroce clades) previously described by Karafas et al. (2015), Wakeman et al. (2015), Leaw et al. (2016), Leung et al. (2017), Lewis et al. (2018), Larsson et al. (2019), Mendes et al. (2019), Nascimento et al. (2019), and Tibiriçá et al. (2020).
Origin of Coolia malayensisConcerning the origin of C. malayensis in the Mediterranean basin, two alternative scenarios can be proposed, introduced versus endemic. Endemicity is difficult to prove since it is not always possible to ascertain whether or not certain species, found once or a few times, have become endemic (Gómez 2006). Moreover, dinoflagellates in the Mediterranean Sea showed a lower percentage of endemic species (Gómez 2006) compared with marine macroscopic species (Bianchi and Morri 2000) or benthic fauna (Fredj et al. 1992).
Initially, C. malayensis was described from the Indo-Pacific region (Leaw et al. 2010) and later Karafas et al. (2015) isolated a C. malayensis strain from the Caribbean and the Gulf Stream in Northwest Atlantic, showing a high dispersal ability. C. malayensis could have been introduced into the Mediterranean Sea either through the Suez Canal or the strait of Gibraltar, or both. The Suez Canal has been considered the major gateway for the entry of invading species from the Red Sea and the Indo-Pacific region (Galil 2009). This could be supported by the fact that the Tunisian strain C. malayensis Com.1 exhibited the lowest p-distance values with C. malayensis CmPD22 (LSU; 0.0055) isolated from the Indian Ocean (Port Dickson, Straits of Malacca) and with CAWD154 (ITS; 0.014) from the south Pacific (Rangaunu Harbour, New Zealand) (Leaw et al. 2016). Further, it has been shown that among dinoflagellates species, most of the Mediterranean Indo-Pacific taxa came from the Pacific Ocean (Gómez 2003a). Indeed, Čalić et al. (2018) suggested that the Suez Canal is the pathway of introduction of the Indo-Pacific planktonic diatoms species Chaetoceros bacteriastroides Karsten and C. pseudosymmetricus. Nevertheless, indication of dinoflagellates species present exclusively in the Mediterranean Sea and Indo-Pacific Ocean are rare (Gómez 2006).
Leaw et al. (2016) hypothesized that C. malayensis was introduced into the Mediterranean Sea during the post-Messinian period by the Zanclean flood when the North Atlantic waters and its biota filled the Mediterranean basin through the strait of Gibraltar. A similar hypothesis has been suggested by Gómez (2003b), to explain the presence of the toxic dinoflagellate Gymnodinium catenatum Graham in the Mediterranean (Gómez et al. 2000). The Atlantic Tunisian current, representing the strongest branch of the Atlantic vein, could also have an important role in this introduction since it flows along the Tunisian coasts and it goes to the south to fuel the flow of the Gulf of Gabès (Ben Ismail et al. 2012, 2014).
Intentional and accidental introductions through ballast waters, ship fouling, aquaculture, trade-in living bait, wrapping of fresh seafood in living algae, aquariology and scientific research could also represent the origin of the C. malayensis introduction to the Mediterranean Sea (Leaw et al. 2016). The importance of the Gulf of Gabès in marine traffic supports this suggestion, as it contains the most important harbor infrastructures in Tunisia (Office de la Marine Marchande et des Ports 2019). For example, Karenia papilionacea and K. bidigitata (Haywood et al. 2004), Gambierdiscus toxicus (Adachi and Fukuyo 1979), and Amphidinium operculatum (Claparède and Lachmann 1859) that were firstly described in the Pacific and Atlantic oceans, respectively, and have since been reported in surrounding harbors and ballast water basins (Dammak-Zouari et al. 2009).
Furthermore, C. malayensis as benthic microalgae is expected to have a more varied pattern of geographical distribution than planktonic species and it might be able to survive dispersal dynamics throughout large basins by passive transport of vegetative and / or resting cysts on floating and drifting objects, such as plastics or wood (Larsson et al. 2019) by water currents, or in the ballast water of ships (Hallegraeff 1993).
Convincingly, the new record of C. malayensis in the South-eastern Mediterranean broadens the geographic distribution pattern of this species and highlights the occurrence of the two closely related taxa C. monotis and C. malayensis in the same area. This contrasts with a previous investigation that suggested that these species do not overlap geographically, with C. monotis showing a colder water distribution (Gómez et al. 2016).
The current study is the first record, supported by both morphological and phylogenetic analyses, of the existence of the benthic dinoflagellate C. malayensis in the Mediterranean Sea. This finding confirmed its circumtropical cosmopolitan distribution and its low degree of endemism and suggests that the Mediterranean C. malayensis may have a Pacific or Atlantic origin. Furthermore, this study demonstrates the co-occurring of C. malayensis with C. monotis. Further investigations using several strains of C. malayensis with broader sampling sites in the Mediterranean basin are required to determine the extent of their distribution and to elucidate their toxicity.
SUPPLEMENTARY MATERIALS
Supplementary Table S1. Sampling sites and dates of Coolia strains isolated from the Gulf of Gabès, Tunisia (https://www.e-algae.org).
Supplementary Table S2. Sampling sites and accession numbers of ITS-5.8S rDNA region and LSU rDNA D1/D3 of strains investigated in this study (https://www.e-algae.org).
ACKNOWLEDGEMENTSWe are grateful to Dr. Isabella Percopo (Electron Microscopy Service, Stazione Zoologica Anton Dohrn) for their assistance in scanning electron microscopy analyses. This work is carried out under the MOBIDOC scheme, funded by the EU through the EMORI program and managed by the ANPR. This study was partially supported by the European funded project “Improving National Capacities in Observation and Management of Marine Environment in Tunisia” (INCOMMET, 295009).
Fig. 2Light micrographs of live Coolia strains. (A & B) C. monotis Com.16. (C & D) C. malayensis Com.1. (E & F) Mucilage aggregates of cells formed during exponential growth of C. monotis Com.10. Scale bars represent: A–D, 20 μm; E, 200 μm; F, 50 μm. 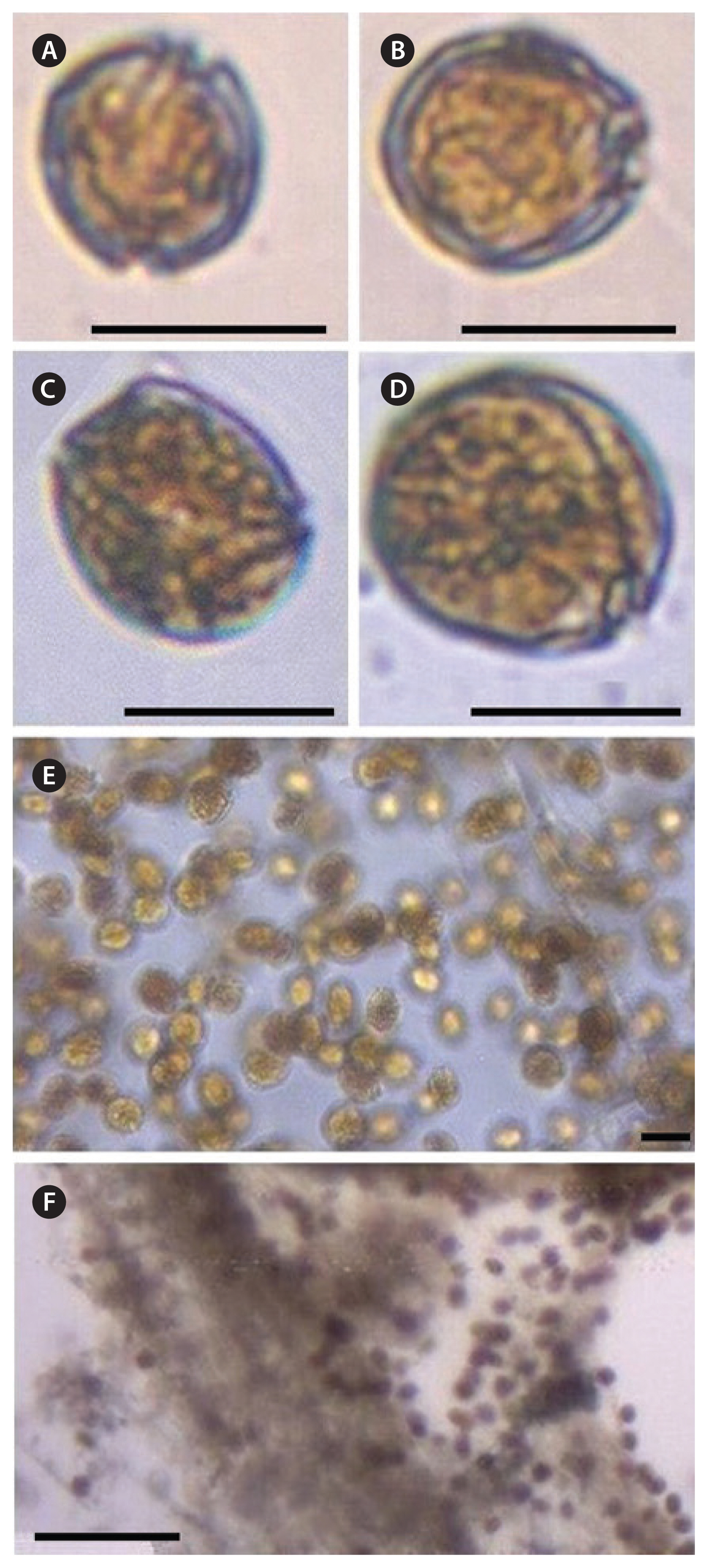
Fig. 3Scanning electron micrographs of Coolia malayensis cells. (A) Apical view of the epithecal plates architecture and the apical pore (Po). (B) Dorsal lateral right side view of hexagonale 1′ and quadrangular 6″ plates. (C) Ventroantapical view of the large 3′′ plate. (D) Antapical view of the hypothecal plates and sulcus (s). (E) Dorsal lateral left side view of the apical pore complex and the narrow cingulum (c). Scale bars represent: A–E, 10 μm. 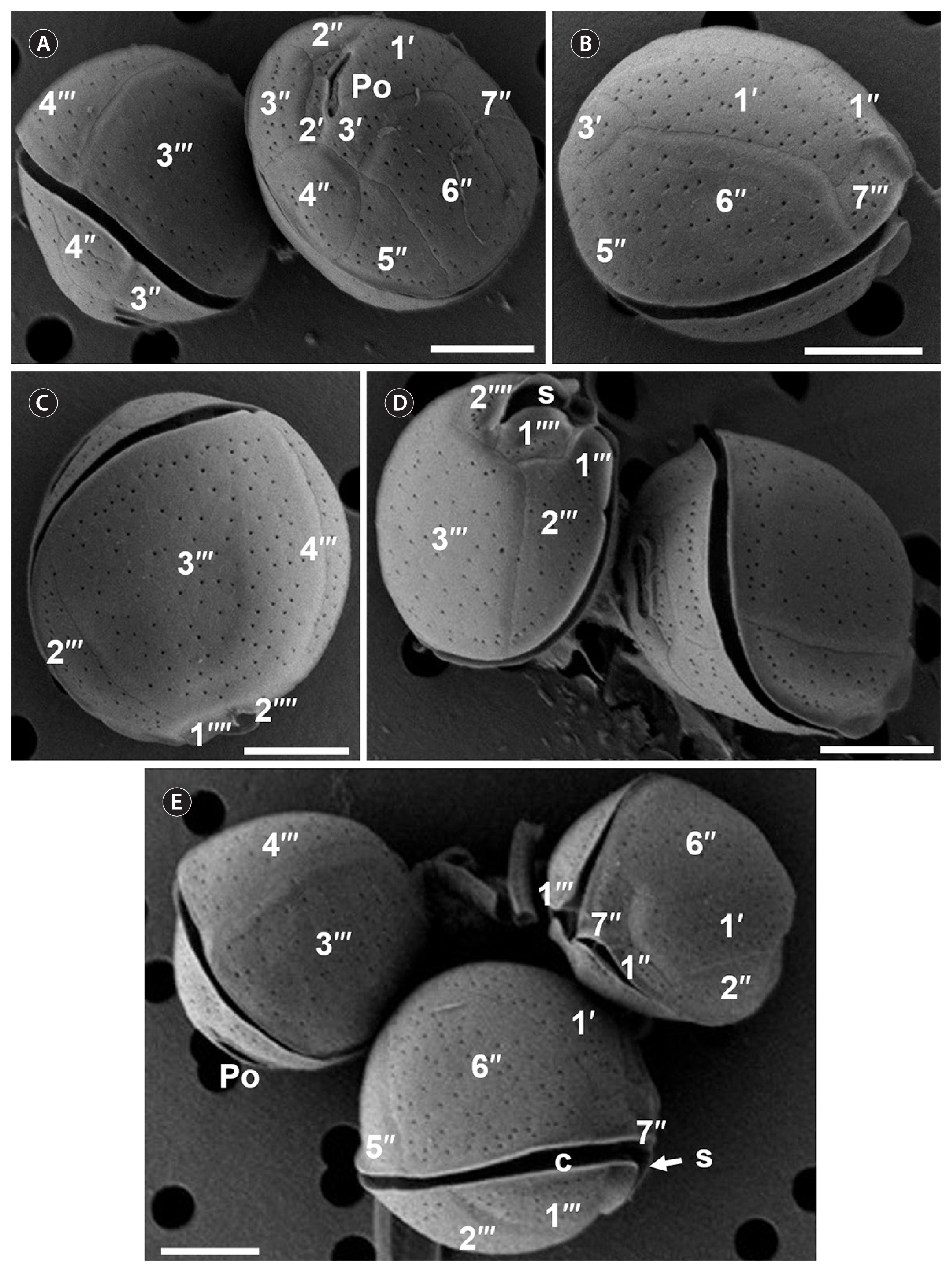
Fig. 4Scanning electron micrographs of Coolia monotis cells. (A & B) Com.10 strain. (A) Cells showing variable size. (B) Apical view of the apical pore (Po), epithecal plates with pores (arrows) on the thecal surface. (C & D) Com.2 strain. (C) Dorsal lateral left side view of hexagonal 1′ plate, apical pore (Po) perforated by row of small pores (arrow) and opening pore (arrowhead). (D) Dorsal lateral right side view of quadrangular 6″ plate. (E & F) Com.6 strain. (E) Ventral view of the epithecal and hypothecal plates, cingulum (c), and sulcus (s). (F) Antapical view of the hypothecal plates. Scale bars represent: A, 100 μm; B, C & E, 1 μm; D & F, 10 μm. 
Fig. 5Transmission electron microscopy images of Coolia malayensis Com.1 (A & E) and C. monotis Com.10 (B–D). (A) Longitudinal section of chloroplasts (chl), nucleus (N), and chromosomes (chr). (B) Transversal sections of nuclear membrane (Nm) with nuclear pores (Np), chromosomes (chr), thylakoids (th), and pyrenoids (py). (C) Rhomboid trichocysts (T) and lipid bodies (lb). (D) Amphiesma with amphiesmal vesicles (Va). (E) Longitudinal sections of spirally coiled fibers (SCF). (F) Golgi body (Gb). Scale bars represent: A, 2 μm; B, D & F, 1 μm; C & E, 0.5 μm. 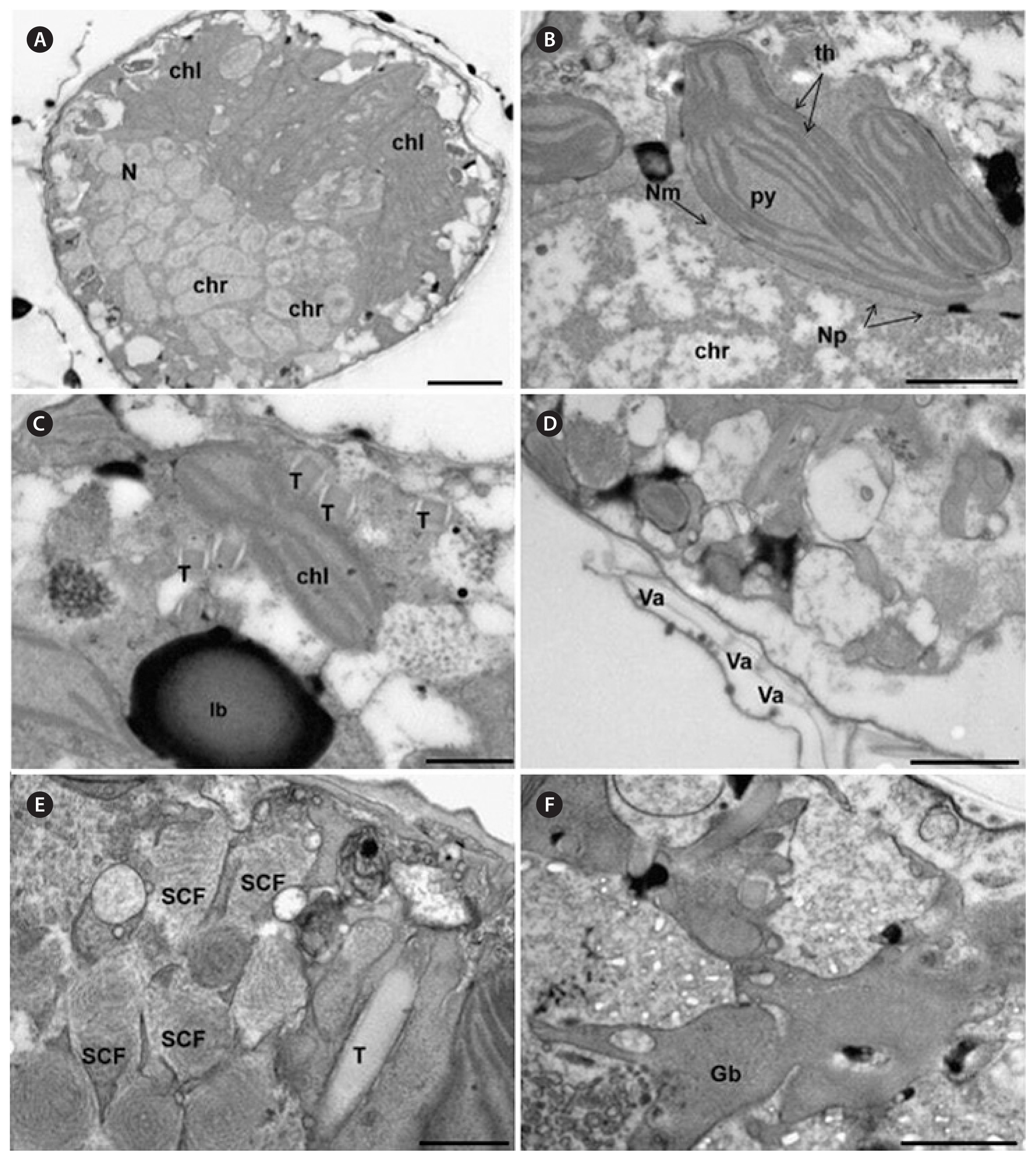
Fig. 6Maximum likelihood (ML) phylogenetic tree of the genus Coolia inferred from the large subunit rDNA gene (D1/D3 region). The tree is rooted with Ostreopsis siamensis (CSIC-D) and Ostreopsis ovata (Oso.3) strains. Each sequence was identified by the GenBank accession number, strain code and isolation site; and it is highlighted according to its geographic area location. Sequences used for the first time in this phylogenetic analysis are in bold. The ML bootstrap values were indicated at nodes, with values lower than 40 are hided. 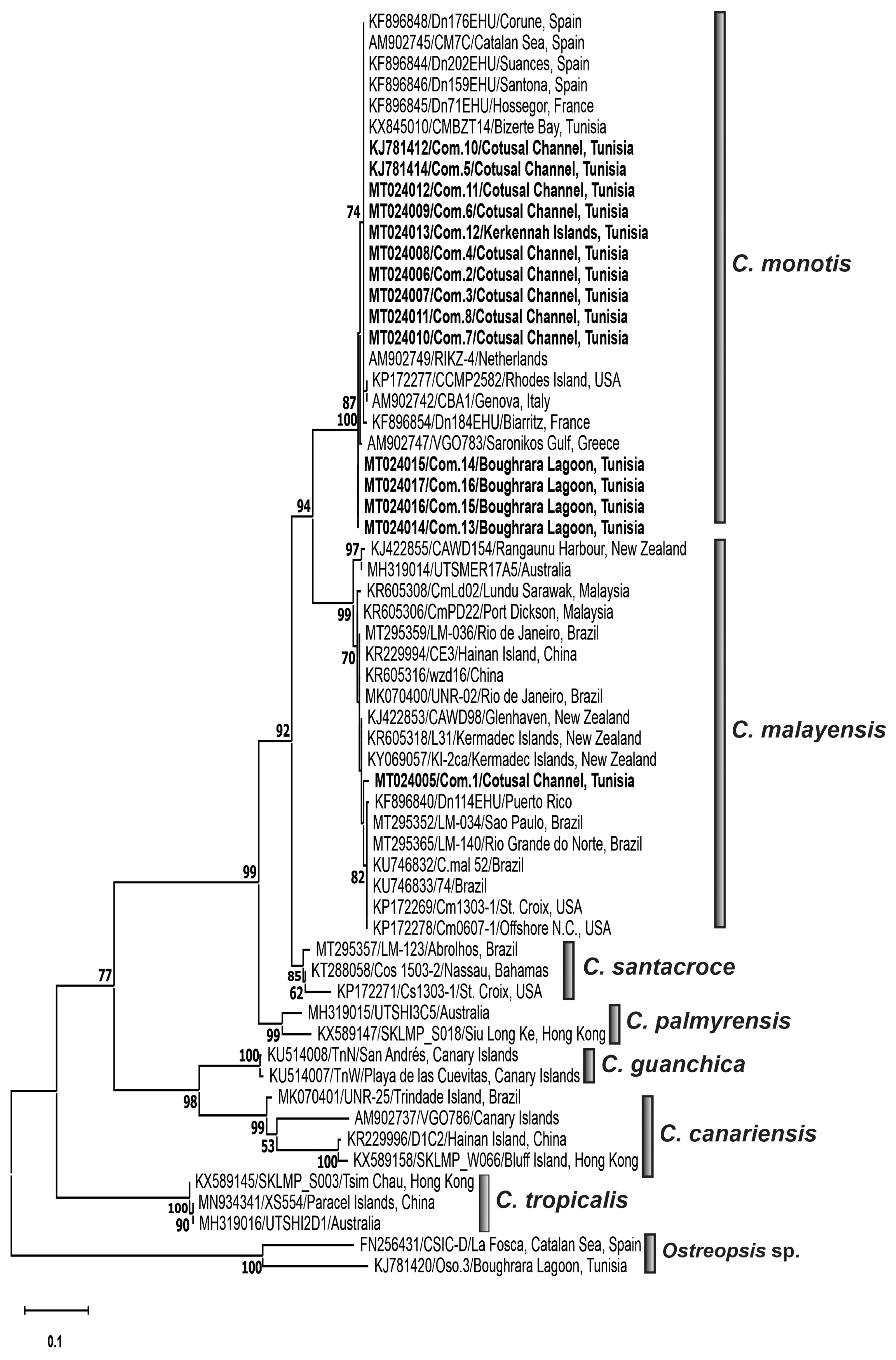
Fig. 7Maximum likelihood (ML) phylogenetic tree of the genus Coolia inferred from the 5.8S rDNA and internal transcribed spacer sequences. The tree is rooted with two strains of Ostreopsis ovata (OVPD7 and Oso.2). Each sequence was identified by the GenBank accession number, strain code and isolation site; and it is highlighted according to its geographic area location. Sequences used for the first time in this phylogenetic analysis are in bold. The ML bootstrap values were indicated at nodes, with values lower than 40 are hided. 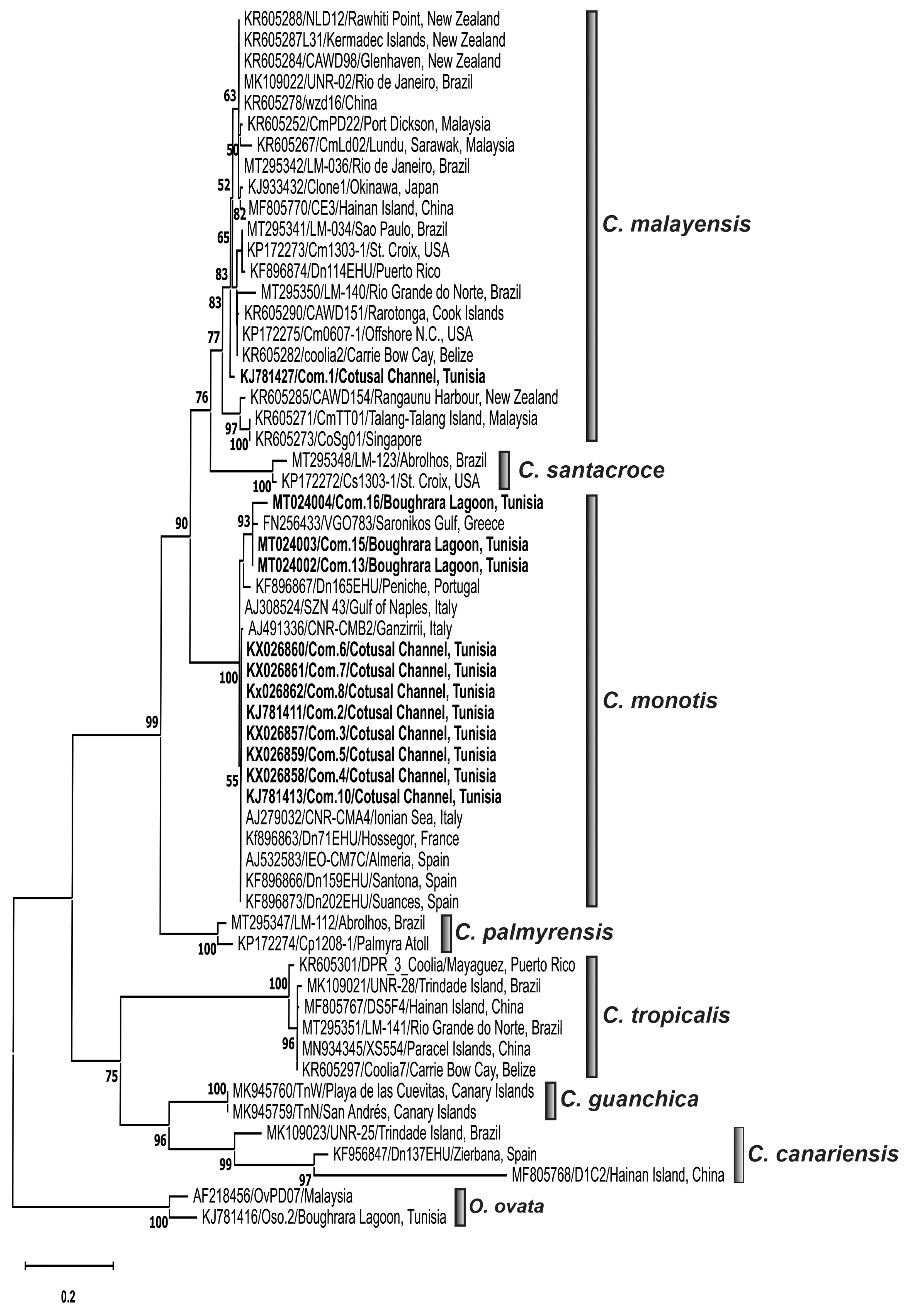
Table 1Morphological features of Coolia malayensis and C. monotis strains isolated from the Gulf of Gabès, Tunisia (South-eastern Mediterranean)
Table 2Evolutionary distances derived from D1–D3 LSU rDNA sequence within and between seven clades of Coolia species Table 3Evolutionary distances derived from ITS-5.8S rDNA region sequence within and between seven clades of Coolia species Table 4Comparison of the morphometric features of Coolia malayensis and C. monotis strains isolated from the Gulf of Gabès with isolates from other areas in the world
REFERENCESAbdennadher, M., Bellaaj Zouari, A., Feki Sahnoun, W., Alverca, E., Penna, A. & Hamza, A. 2017.
Ostreopsis cf. ovata in the Gulf of Gabès (south-eastern Mediterranean Sea): morphological, molecular and ecological characterization. Harmful Algae. 63:56–67.
Abdennadher, M., Bellaaj Zouari, A., Feki Sahnoun, W., Dammak Walha, L., Mahfoudi, M. & Hamza, A. 2020. A long-term study on Coolia monotis distribution from the South-east. Mediterranean Sea Cont Shelf Res. 211:104267 pp.
Adachi, M., Sako, Y. & Ishida, Y. 1994. Restriction fragment length polymorphism of ribosomal DNA internal transcribed spacer and 5.8S regions in Japanese Alexandrium species (Dinophyceae). J Phycol. 30:857–865.
Adachi, R. & Fukuyo, Y. 1979. The thecal structure of a marine toxic dinoflagellate Gambierdiscus toxicus gen. et sp. nov. collected in a ciguatera-endemic area. Bull Jpn Soc Sci Fish. 45:67–71.
Aligizaki, K. & Nikolaidis, G. 2006. The presence of the potentially toxic genera Ostreopsis and Coolia (Dinophyceae) in the north Aegean Sea, Greece. Harmful Algae. 5:717–730.
Al-Yamani, FY. & Saburova, MA. 2010. Illustrated guide on the flagellates of Kuwait’s intertidal soft sediments. Kuwait Institute for Scientific Research, Safat, 197 pp.
Andersen, RA. 2005. Algal culturing techniques. Elsevier Academic Press, Burlington, MA, 565 pp.
Armi, Z., Turki, S., Trabelsi, E. & Maiz, NB. 2010. First recorded proliferation of Coolia monotis (Meunier, 1919) in the North Lake of Tunis (Tunisia) correlation with environmental factors. Environ Monit Assess. 164:423–433.
Balech, E. 1956. Etude des Dinoflagelles du sable de Roscoff. Rev Algol. 2:29–52.
Ben-Gharbia, H., Yahia, OK-D., Amzil, Z., Chomérat, N., Abadie, E., Masseret, E., Sibat, M., Triki, HZ., Nouri, H. & Laabir, M. 2016. Toxicity and growth assessments of three thermophilic benthic dinoflagellates (Ostreopsis cf. ovata, Prorocentrum lima and Coolia monotis) developing in the southern Mediterranean basin. Toxins. 8:297 pp.
Ben Ismail, S., Sammari, C., Gasparini, GP., Béranger, K., Brahim, M. & Aleya, L. 2012. Water masses exchanged through the Channel of Sicily: evidence for the presence of new water masses on the Tunisian side of the channel. Deep Sea Res I Oceanogr Res PapI. 63:65–81.
Ben Ismail, S., Schroeder, K., Sammari, C., Gasparini, GP., Borghinic, M. & Aleya, L. 2014. Interannual variability of water mass properties in the Tunisia-Sicily Channel. J Mar Syst. 135:14–28.
Besada, EG., Loeblich, LA. & Loeblich, AR. 3rd 1982. Observations on tropical, benthic dinoflagellates from ciguatera-endemic areas: Coolia, Gambierdiscus, and Ostreopsis
. Bull Mar Sci. 32:723–735.
Bianchi, CN. & Morri, C. 2000. Marine biodiversity of the Mediterranean Sea: situation, problems and prospects for future research. Mar Pollut Bull. 40:367–376.
Čalić, M., Ljubimir, S., Bosak, S. & Car, A. 2018. First records of two planktonic Indo-Pacific diatoms: Chaetoceros bacteriastroides and C. pseudosymmetricus in the Adriatic Sea. Oceanologia. 60:101–105.
Claparède, É. & Lachmann, J. 1859. Études sur les infusoires et les rhizopodes. Mém l’Inst Natl Genev. 6:261–482.
Dammak-Zouari, H., Hamza, A. & Bouain, A. 2009. Gymnodiniales in the Gulf of Gabes (Tunisia). Cah Biol Mar. 50:153–170.
David, H., Laza-Martínez, A., Miguel, I. & Orive, E. 2014. Broad distribution of Coolia monotis and restricted distribution of Coolia cf. canariensis (Dinophyceae) in the Atlantic coast of the Iberian Peninsula. Phycologia. 53:342–352.
David, H., Laza-Martínez, A., Rodríguez, F., Fraga, S. & Orive, E. 2019.
Coolia guanchica sp. nov. (Dinophyceae) a new epibenthic dinoflagellate from the Canary Islands (NE Atlantic Ocean). Eur J Phycol. 55:76–88.
Dolapsakis, NP., Kilpatrick, MW., Economou-Amilli, A. & Tafas, T. 2006. Morphology and rDNA phylogeny of a Mediterranean Coolia monotis (Dinophyceae) strain from Greece. Sci Mar. 70:67–76.
Faust, MA. 1995. Observation of sand-dwelling toxic dinoflagellates (Dinophyceae) from widely differing sites, including two new species. J Phycol. 31:996–1003.
Faust, MA. 2009. Ciguatera-causing dinoflagellates in a coral-reef mangrove ecosystem, Belize. Atoll Res Bull. 569:1–32.
Felsenstein, J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 17:368–376.
Fraga, S., Penna, A., Bianconi, I., Paz, B. & Zapata, M. 2008.
Coolia canariensis sp. nov. (Dinophyceae), a new nontoxic epiphytic benthic dinoflagellate from the Canary Islands. J Phycol. 44:1060–1070.
Fredj, G., Bellan-Santin, D. & Meinardi, M. 1992. Etat des connaissances sur la faune marine méditerranéenne. Bull l’Inst Océanogr Monaco. 9:133–145.
Galil, BS. 2009. Taking stock: inventory of alien species in the Mediterranean Sea. Biol Invasions. 11:359–372.
Giussani, V., Sbrana, F., Asnaghi, V., Vassalli, M., Faimali, M., Casabianca, S., Penna, A., Ciminiello, P., Dell’Aversano, C., Tartaglione, L., Mazzeo, A. & Chiantore, M. 2015. Active role of the mucilage in the toxicity mechanism of the harmful benthic dinoflagellate Ostreopsis cf. ovata
. Harmful Algae. 44:46–53.
Gómez, F. 2003b. The toxic dinoflagellate Gymnodinium catenatum: an invader in the Mediterranean Sea. Acta Bot Croat. 62:65–72.
Gómez, F. 2006. Endemic and Indo-Pacific plankton in the Mediterranean Sea: a study based on dinoflagellate records. J Biogeogr. 33:261–270.
Gómez, F., Echevarria, F., García, CM., Prieto, L., Ruiz, J., Reul, A., Jiménez-Gómez, F. & Varela, M. 2000. Micro- plankton distribution in the Strait of Gibraltar: coupling between organisms and hydrodynamic structures. J Plankton Res. 22:603–617.
Gómez, F., Qiu, D., Otero-Morales, E., Lopes, RM. & Lin, S. 2016. Circumtropical distribution of the epiphytic dinoflagellate Coolia malayensis (Dinophyceae): morphology and molecular phylogeny from Puerto Rico and Brazil. Phycol Res. 64:194–199.
Guillard, RRL. & Hargraves, PE. 1993.
Stichochrysis inmobilis is a diatom, not a chrysophyte. Phycologia. 32:234–236.
Halim, Y. 1960. Etude quantitative et qualitative du cycle écologique des Dinoflagellés dans les eaux de Villefranche-sur-Mer. Ann Inst Océanogr. 38:123–232.
Hallegraeff, GM. 1993. A review of harmful algal blooms and their apparent global increase. Phycologia. 32:79–99.
Hattour, A. & Ben Mustapha, K. 2013. Le couvert végétal marin du golfe de Gabès: Cartographie et réseau de surveillance de l’herbier de Posidonie. Institut National des Sciences et Technologies de la Mer, Salammbô, 164 pp.
Haywood, AJ., Steidinger, KA., Truby, EW., Bergquist, PR., Bergquist, PL., Adamson, J. & MacKenzie, L. 2004. Comparative morphology and molecular phylogenetic analysis of three new species of the genus Karenia (Dinophyceae) from New Zealand. J Phycol. 40:165–179.
Ho, TV. & Nguyen, LN. 2014. Morphology and distribution of the three epiphytic dinoflagellate species Coolia monotis, C. tropicalis, and C. canariensis (Ostreopsidaceae, Gonyaulacales, Dinophyceae) from Vietnamese coastal waters. Ocean Sci J. 49:211–221.
Holmes, MJ., Lewis, RJ., Jones, A. & Hoy, AW. 1995. Cooliatoxin, the first toxin from Coolia monotis (Dinophyceae). Nat. Toxins. 3:355–362.
Honsell, G., Bonifacio, A., De Bortoli, M., Penna, A., Battocchi, C., Ciminiello, P., Dell’Aversano, C., Fattorusso, E., Sosa, S., Yasumoto, T. & Tubaro, A. 2013. New insights on cytological and metabolic features of Ostreopsis cf. ovata Fukuyo (Dinophyceae): a multidisciplinary approach. PLoS ONE. 8:e57291 pp.
Ismael, AA. 2014. First record of Coolia monotis Meunier along Alexandria coast: Egypt. Egypt J Aquat Res. 40:19–25.
Karafas, S., York, R. & Tomas, C. 2015. Morphological and genetic analysis of the Coolia monotis species complex with the introduction of two new species, Coolia santacroce sp. nov. and Coolia palmyrensis sp. nov. (Dinophyceae). Harmful Algae. 46:18–33.
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
Larsson, ME., Smith, KF. & Doblin, MA. 2019. First description of the environmental niche of the epibenthic dinoflagellate species Coolia palmyrensis, C. malayensis, and C. tropicalis (Dinophyceae) from Eastern Australia. J Phycol. 55:565–577.
Laza-Martínez, A., Orive, E. & Miguel, I. 2011. Morphological and genetic characterization of benthic dinoflagellates of the genera Coolia, Ostreopsis, and Prorocentrum from the south-eastern Bay of Biscay. Eur J Phycol. 46:45–65.
Leaw, CP., Lim, PT., Ahmad, A. & Usup, G. 2001. Genetic diversity of Ostreopsis ovata (Dinophyceae) from Malaysia. Mar Biotechnol. 3:246–255.
Leaw, C-P., Lim, P-T., Chen, K-W., Ng, B-K. & Usup, G. 2010. Morphology and molecular characterization of a new species of thecate benthic dinoflagellate, Coolia malayensis sp. nov. (Dinophyceae). J Phycol. 46:162–171.
Leaw, CP., Tan, TH., Lim, HC., Teng, ST., Yong, HL., Smith, KF., Rhodes, L., Wolf, M., Holland, WC., Vandersea, MW., Litaker, RW., Tester, PA., Gu, H., Usup, G. & Lim, PT. 2016. New scenario for speciation in the benthic dinoflagellate genus Coolia (Dinophyceae). Harmful Algae. 55:137–149.
Leung, PTY., Yan, M., Yiu, SKF., Lam, VTT., Ip, JCH., Au, MWY., Chen, C-Y., Wai, T-C. & Lam, PKS. 2017. Molecular phylogeny and toxicity of harmful benthic dinoflagellates Coolia (Ostreopsidaceae, Dinophyceae) in a sub-tropical marine ecosystem: the first record from Hong Kong. Mar Pollut Bull. 124:878–889.
Lewis, NI., Wolny, JL., Achenbach, JC., Ellis, L., Pitula, JS., Rafuse, C., Rosales, DS. & McCarron, P. 2018. Identification, growth and toxicity assessment of Coolia Meunier (Dinophyceae) from Nova Scotia, Canada. Harmful Algae. 75:45–56.
Loukil-Baklouti, A., Feki-Sahnoun, W., Hamza, A., Abdennadher, M., Mahfoudhi, M., Bouain, A. & Jarboui, O. 2018. Controlling factors of harmful microalgae distribution in water column, biofilm and sediment in shellfish production area (South of Sfax, Gulf of Gabes) from southern Tunisia. Cont Shelf Res. 152:61–70.
Madeira, F., Park, YM., Lee, J., Buso, N., Gur, T., Madhusoodanan, N., Basutkar, P., Tivey, ARN., Potter, SC., Finn, RD. & Lopez, R. 2019. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47:W636–W641.
Mendes, MCQ., Nunes, JMC., Fraga, S., Rodríguez, F., Franco, JM., Riobó, P., Branco, S. & Menezes, M. 2019. Morphology, molecular phylogeny and toxinology of Coolia and Prorocentrum strains isolated from the tropical South Western Atlantic Ocean. Bot Mar. 62:125–140.
Meunier, A. 1919. Microplankton de la Mer Flamande. III Les Peridiniens. Mem Mus R Hist Nat Belg. 8:116–144.
Mikulski, CM., Morton, SL. & Doucette, GJ. 2005. Development and application of LSU rRNA probes for Karenia brevis in the Gulf of Mexico, USA. Harmful Algae. 4:49–60.
Mohammad Noor, N., Adam, A., Saad, S., Khodzori, FA. & Shaleh, SRM. 2019. Substrate preference and effects of medium with soil extract on growth of genus Coolia (Dinophyceae). J Sustain Sci Manag. 14:71–81.
Mohammad-Noor, N., Moestrup, Ø., Lundholm, N., Fraga, S., Adam, A., Holmes, MJ. & Saleh, E. 2013. Autecology and phylogeny of Coolia tropicalis and Coolia malayensis (Dinophyceae), with emphasis on taxonomy of C. tropicalis based on light microscopy, scanning electron microscopy and LSU rDNA. J Phycol. 49:536–545.
Momigliano, P., Sparrow, L., Blair, D. & Heimann, K. 2013. The diversity of Coolia spp. (Dinophyceae Ostreopsidaceae) in the Central Great Barrier Reef Region. PLoS ONE. 8:e79278 pp.
Moncer, M., Hamza, A., Feki-Sahnoun, W., Mabrouk, L. & Bel Hassen, M. 2017. Variability patterns of epibenthic microalgae in eastern Tunisian coasts. Sci Mar. 81:487–498.
Nascimento, SM., da Silva, RAF., Oliveira, F., Fraga, S. & Salgueiro, F. 2019. Morphology and molecular phylogeny of Coolia tropicalis, Coolia malayensis and a new lineage of the Coolia canariensis species complex (Dinophyceae) isolated from Brazil. Eur J Phycol. 54:484–496.
Nguyen, NL. & Larsen, J. 2004. Gonyaulacales. In : Larsen J, Nguyen NL, editors Guide to the Identification of Potentially Toxic Microalgae in Vietnamese Waters. Opera Bot. 140. Copenhagen, 73–116.
Office de la Marine Marchande et des Ports 2019. Rapport annuel. Office de la Marine Marchande et des Ports, La Goulette, 46 pp.
Pagliara, P. & Caroppo, C. 2012. Toxicity assessment of Amphidinium carterae, Coolia cfr. monotis and Ostreopsis cfr. ovata (Dinophyta) isolated from the northern Ionian Sea (Mediterranean Sea). Toxicon. 60:1203–1214.
Penna, A., Vila, M., Fraga, S., Giacobbe, MG., Andreoni, F., Riobó, P. & Vernesi, C. 2005. Characterization of Ostreopsis and Coolia (Dinophyceae) isolates in the western Mediterranean Sea based on morphology, toxicity and internal transcribed spacer 5.8S rDNA sequences. J Phycol. 41:212–225.
Reynolds, CS. 2007. Variability in the provision and function of mucilage in phytoplankton: facultative responses to the environment. Hydrobiologia. 578:37–45.
Rhodes, L., Smith, K., Harwood, T. & Bedford, C. 2014a. Novel and toxin-producing epiphytic dinoflagellates isolated from sub-tropical Raoul Island, Kermadec Islands group. N Z J Mar Freshw Res. 48:594–599.
Rhodes, L., Smith, K., Papiol, GG., Adamson, J., Harwood, T. & Munday, R. 2014b. Epiphytic dinoflagellates in sub-tropical New Zealand, in particular the genus Coolia Meuneir. Harmful Algae. 34:36–41.
Rhodes, LL. & Thomas, AE. 1997.
Coolia monotis (Dinophyceae): a toxic epiphytic microalgal species found in New Zealand (note). N Z J Mar Freshw Res. 31:139–141.
Scholin, CA., Herzog, M., Sogin, M. & Anderson, DM. 1994. Identification of group and strain-specific genetic markers for globally distributed Alexandrium (Dinophyceae). II. Sequence analysis of a fragment of the LSU rRNA gene. J Phycol. 30:999–1011.
Shah, MMR., Samarakoon, KW., Ko, J-Y., Chaminda Lakmal, HH., Lee, J-H., An, S-J., Jeon, Y-J. & Lee, J-B. 2014. Potentiality of benthic dinoflagellate cultures and screening of their bioactivities in Jeju Island, Korea. Afr J Biotechnol. 13:792–805.
Tamura, K. 1992. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol Biol Evol. 9:678–687.
Tamura, K. & Nei, M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 10:512–526.
Tawong, W., Nishimura, T., Sakanari, H., Sato, S., Yamaguchi, H. & Adachi, M. 2015. Characterization of Gambierdiscus and Coolia (Dinophyceae) isolates from Thailand based on morphology and phylogeny. Phycol Res. 63:125–133.
Ten-Hage, L., Turquet, J., Quod, JP. & Couté, A. 2000.
Coolia areolata sp. nov. (Dinophyceae), a new sand-dwelling dinoflagellate from the southwestern Indian Ocean. Phycologia. 39:377–383.
Tibiriçá, CEJA., Sibat, M., Fernandes, LF., Bilien, G., Chomérat, N., Hess, P. & Mafra, LL. Jr 2020. Diversity and toxicity of the genus Coolia Meunier in Brazil, and detection of 44-methyl gambierone in Coolia tropicalis
. Toxins. 12:327 pp.
Vila, M., Garcés, E. & Masó, M. 2001. Potentially epiphytic dinoflagellate assemblages on macroalgae in the NW Mediterranean. Aquat Microbiol Ecol. 26:51–60.
Wakeman, KC., Yamaguchi, A., Roy, MC. & Jenke-Kodama, H. 2015. Morphology, phylogeny and novel chemical compounds from Coolia malayensis (Dinophyceae) from Okinawa, Japan. Harmful Algae. 44:8–19.
Zhang, H., Lü, S., Cen, J., Li, Y., Li, Q. & Wu, Z. 2020. Morphology and molecular phylogeny of three species of Coolia (Dinophyceae) from Hainan Island, South China Sea. J Oceanol Limnol, Advanced publication,
https://doi.org/10.1007/s00343-020-9326-z
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||