ABSTRACTGenetic study of haploid organisms offers the advantage that mutant phenotypes are directly displayed, but has the disadvantage that strains carrying lethal mutations are not readily maintained. We describe an approach for generating and performing genetic analysis of diploid strains of Chlamydomonas reinhardtii, which is normally haploid. First protocol utilizes self-mating diploid strains that will facilitate the genetic analysis of recessive lethal mutations by offering a convenient way to produce homozygous diploids in a single mating. Second protocol is designed to reduce the chance of contamination and the accumulation of spontaneous mutations for long-term storage of mutant strains. Third protocol for inducing the meiotic program is also included to produce haploid mutant strains following tetraploid genetic analysis. We discuss implication of self-fertile strains for the future of Chlamydomonas research.
INTRODUCTIONGenetic analysis of Chlamydomonas reinhardtii has largely focused on two traits_photosynthesis and flagellar motility / assembly_in part because neither trait exists in yeast and in part because neither process is essential for viability: photosynthetic mutants survive with acetate supplementation, and flagella are not necessary for growth.
With the completion of the C. reinhardtii genome, a cornucopia of new genes awaits functional and interactional analysis (Merchant et al. 2007, Blaby et al. 2014, Romero-Campero et al. 2016). However, unlike the photosynthetic and flagellar gene complement, mutations in many of these genes are likely to be lethal in haploids. Recently a large-scale insertional mutant library became available, in which ~37,000 individual mutations are mapped on the genome and cover 73% of all annotated genes (Li et al. 2016). When excluding about 20% of the insertions in 3′ untranslated region as benign mutations not causing defects, ~50% of the genes lack a loss-of-function mutation, indicating the need for additional methods to study a large number of essential genes whose mutation in haploids result in lethality.
One way to bypass the lethality of essential genes in haploids is to induce and maintain the mutations in diploids and then screen for their effects either in haploid meiotic progeny or in diploids amenable to genetic analysis. To date, diploid strains of C. reinhardtii have largely been used to assess dominance and complementation relationships using so-called heterozygous diploids. In the original protocol (Ebersold 1967), mutant χ is first crossed to an arg7 strain and χ arg7 progeny are recovered; χ arg7 is then mated with an arg2 strain, and the mating mixture is plated on an arginine-free medium. Most mated cells form zygotes, but a highly variable minority (reported range, 0.08% [S. Dutcher personal communication] to 4% [Ebersold 1967]) instead resume mitotic growth as stable diploids, where arg2/arg7 complementation allows auxotrophic growth. The dominant / recessive nature of χ is then assessed in these diploids, and introduction of alternative χ alleles via the arg2 parent allows assessment of their complementarity (Fig. 1B). Additional selection schemes for heterozygous diploid recovery have more recently been described (Palombella and Dutcher 1998, Bellafiore et al. 2002).
A major drawback of the heterozygous diploids is that they are refractory to genetic analysis. When induced to undergo gametogenesis by nitrogen starvation, they invariably differentiate as minus gametes due to their possession of the dominant MID gene (Ferris and Goodenough 1997) in the mt− locus, and hence can only mate with plus gametes. Matings with haploid plus gametes generate triploid zygotes that undergo abortive meioses. To avoid the abortive triploid meiosis, Dutcher reported a method using drugs affecting microtubule stability that prevented nuclear fusion during mating and therefore allowed successful meiosis of the diploid nucleus from a mating between haploid and diploid strains (Dutcher 1988). Investigators have also developed techniques for generating diploid plus strains using polyethylene glycol (PEG)-mediated protoplast cell fusion (Matagne et al. 1979, Galloway and Holden 1984), followed by tetraploid genetics (Galloway and Goodenough 1985) (Fig. 1C). Several diploid plus strains currently available in the Chlamydomonas Resource Center were generated by the PEG method. However, the PEG method is inefficient and finicky, with reports of chromosome loss in yeast (Molnar and Sipiczki 1993), and the diploid products of tetraploid meioses fail to express recessive traits without additional backcrosses.
During the course of a study elucidating the homeoprotein-based induction of haploid → diploid → meiosis transitions in C. reinhardtii, we have developed procedures that generate diploid cell lines with the capacity to undergo self-mating or to undergo meiosis without mating. These techniques and strains were briefly described in a previous publication (Lee et al. 2008) in the context of analyzing homeoprotein control of zygote development. Their description is expanded here to offer novel resources to benefit future genetic studies of C. reinhardtii, where we lift up their general applicability and note some additional usages.
MATERIALS AND METHODSStrains and culture conditions
C. reinhardtii strains CC-2926 (mt−, iso1[T-ARG7], arg7, also known as isolr:pARG7.8), CC-124 (mt−, AC17 nit2), and CC-125 (mt+, AC17 nit2) are available from the Chlamydomonas Resource Center (https://www.chlamycollection.org/). The ac17 NIT2 strains used in vegetative diploids were obtained from Dutcher laboratory (Washington Univ.). All C. reinhardtii strains were maintained and cultured under medium light (50 μmol photons m−2 s−1) at 23°C on solid (1.5% agar) Tris-acetate-phosphate (TAP) medium plates (Harris 1989). Nitrogen-free TAP (N-free TAP) medium was prepared by omitting nitrogen.
Gametogenesis, mating and zygote isolationPreparation of cells for gametogenesis, mating, and zygote / tetrad isolation were performed as described (Lee et al. 2008). Gametogenesis samples were harvested from cultures grown on TAP plates for 5–7 days, washed once and resuspended in N-free TAP to a final 5 × 107 cells mL−1. After 4 to 6 h, the cells in N-free TAP displayed agglutination when mixed with tester gametes of opposite mating type, the indication of gametogenesis. Zygotes for tetrad isolation were prepared from a mating mixture of an equal number of plus and minus gametes that were plated on N-free TAP 4% agar plates, incubated in the light for 1 day and the dark for more than 5 days. These mature zygotes were transferred to TAP medium, then germinated and produced 4 to 16 progeny per zygote in 12–24 h after transfer to the light.
Generation of vegetative diploidsDiploids were generated by the method described in Palombella and Dutcher (1998). Two hundred fifty microliters of active gamete preparations (107 cells mL−1) from haploid parental strains carrying ac17 NIT2 or AC17 nit2 were mixed for mating and incubated for 2 h when small cell clumps become visible. The mating mixture was then plated on the selection plate containing TP medium (TAP omitting acetate) with 2 mM sodium nitrate as the sole nitrogen source. Colonies were visible after 10 days of growth. Diploidy was confirmed by polymerase chain reaction (PCR)-based determination of heterozygous mating types and eventually by cellular DNA content. Primers used in genotyping are provided in Table 1.
Determination of DNA contentEach strain was clonally isolated on agar plates before analysis. Cells were grown to stationary phase (>107 cells mL−1) and maintained at least 24 h to assure all possessed a 1C DNA content, and total DNA was extracted from 1 mL aliquots as described (Gallaher et al. 2015). Interphase blocker (Phase Lock Gel; QuantaBio, Beverly, MA, USA) was used to maximize recovery of the aqueous phase during phenol / chloroform extraction. Extracted DNA was treated with RNase A for 3 min at 37°C and precipitated with 4 M sodium chloride, 20% PEG-8000 before being analyzed by DNA-specific fluorometry using Qubit dsDNA HS assay kit (Q32851; Invitrogen, Carlsbad, CA, USA).
Estimation of cell volumeMid-logarithmic cultures were harvested to capture cells in all cell cycle stages. Only cells without cleavage furrows were analyzed. Individual areas of more than 95 cells were measured using ImageJ from micrographs taken on a hemocytometer, expressed as μm2. Area to volume conversion was done by Eq. (1).
Desiccation protocolEach strain was grown on TAP-agar plate for a week and transferred to liquid N-free TAP medium for 24 h before harvest; the products of WT matings were harvested at 2 h after mixing plus and minus gametes. One point five milliliters of harvested culture (107 cell mL−1) was added to an equal volume of adsorbent clay particles aliquoted in sterile glass tubes. Following storage in the dark for the indicated times, a small green sector of the particle / cell mixture was transferred to 2 mL of TAP medium, and survival was examined after one week. Cat litter from natural clay (Dr. Elsey’s “Ultra Precious Cat” brand) was used as the adsorbent.
RESULTS AND DISCUSSIONWe have developed two novel approaches: the use of the iso1 mutant strain to generate diploids that are homozygous for mating-type and polyploids, and the use of homeoproteins to drive such diploids into meiosis. We also developed strain storage protocols. These are described in turn (summarized in Fig. 1D–F).
Self-mating diploid generation using the mutation iso1iso1 mutationThe iso1 mt− strain carries a stable insertional mutation that generates an atypical gametogenesis: some cells in a clone differentiate as minus gametes and the rest as plus gametes, the result being intraclonal isoagglutination (Campbell et al. 1995). Campbell et al. (1995) document that each cell expresses either plus or minus agglutinins and not a mixture. The isoagglutinating cells do not fuse to form zygotes, however, because the plus gametes lack the FUS1 gene that is located in the mt+ locus and required for plus-minus gametic fusion.
The iso1 mutation, an insertion in the gene encoding the chromatin-remodeling protein ISWI (Lin 2006), was reported to be recessive in heterozygous diploids generated from an iso1 arg2 mt− strain (Campbell et al. 1995). However, Lin (2006) was unable to complement the iso1 phenotype with the wild type copy of ISO1. To explore this discrepancy, we constructed diploids using an ISO1 ac17 NIT2 mt+ × iso1 AC17 nit2 mt− mating and the selection scheme described by Palombella and Dutcher (1998), and were able to recover iso1/ISO1 mt+/mt− heterozygous diploids that display the isoagglutination phenotype (Table 2).
To further evaluate this apparent dominance of iso1, we performed reciprocal matings — iso1 AC17 nit2 mt+ × ISO1 ac17 NIT2 mt− and ISO1 ac17 NIT2 mt+ × iso1 AC17 nit2 mt− — and homozygous matings — iso1 ac17 NIT2 mt+ × iso1 AC17 nit2 mt− — and collected vegetative diploids (Table 2).
All the iso1 homozygous diploids showed strong and stable isoagglutination. None of the ISO1/iso1 heterozygous diploids receiving iso1 from a plus parent showed isoagglutination, whereas heterozygous ISO1/iso1 diploids receiving iso1 from a minus parent initially showed isoagglutination but this gradually weakened and displayed minus phenotype over months in culture. These results cannot be explained by simple dominant or recessive behavior of the iso1 allele. Instead, the isoagglutination phenotype may possess an inherent, if transient, stability even in a heterozygous ISO1/iso1 background but not readily develop de novo in a heterozygous background.
Given these observations, we were unable to ascertain whether the iso1 mutation is recessive or dominant. Nonetheless, the unusual behavior of iso1, and its location in the ISWI chromatin-remodeling gene, suggest possible epigenetic regulation of sexual phenotypes. For example, the full expression of MID and / or MTD1, two genes encoded exclusively in the MT− locus, may be facilitated by the Iso1 protein, a hypothesis supported by the isoagglutination phenotype of plus strains harboring a transgenic MID gene which may undergo variable regulation in its transgenic location in the absence of MTD1 (Ferris and Goodenough 1997, Lin and Goodenough 2007).
The FUS1 gene escapes MID repression in heterozygous diploids (Ferris et al. 1996). Hence the subset of iso1 diploid gametes behaving as plus are capable of fusing with the iso1 diploid gametes behaving as minus to generate tetraploid zygotes which form a zygotic pellicle matrix, the hallmark of successful zygote development (Lee et al. 2008).
Benefits of selfing diploidsThe intraclonal mating or selfing capability of iso1 diploids allows direct generation of tetraploid zygotes that yield diploid meiotic progeny heterozygous or homozygous for genes of interest (Fig. 1D). Analysis of the heterozygotes allows assessment of dominant / recessive relationships, while analysis of the homozygotes allows assessment of mutant phenotypes. The collection of homozygous diploids would normally take two generations of crosses and multiple rounds of genotyping using mt+/mt+ and mt+/mt− diploids.
Pan et al. (2004) have reviewed the advantages of diploid lines for yeast analysis. At present, Chlamydomonas mutant screens usually entail forward genetics: insertional mutagenesis of haploid strains is followed by time-consuming screens for phenotypes of interest and identification of affected genes by their linkage to insertion sequences or directly by whole-genome-sequencing (e.g., Dent et al. 2015, Lin et al. 2018). Despite technical advances in mutant identification, the use of haploid strains disallows recovery of haploid-lethal mutants. Our selfing diploid strains provide a solution to collection and analysis of mutations in essential genes in C. reinhardtii.
Polyploid geneticsIn a previous study, tetraploid genetics was carried out using PEG-mediated diploids as parents (Fig. 1C) (Galloway and Goodenough 1985). To establish tetraploid genetics protocol that does not involve PEG, we carried out genetic analysis of heterozygous diploids carrying iso1 that differentiate into both plus and minus diploid gametes (Fig. 1D). We first tested whether the iso1 mutation behaves as dominant or recessive in the meiotic progeny. Germination of the self-mated tetraploid zygotes yielded ISO1/iso1 diploids in mt+/mt− and mt−/mt− backgrounds. All the heterozygous iso1 progeny possess at least one mt− locus, and all showed strong isoagglutination, confirming that iso1 mutation is indeed dominant. This conclusion, which contrasts with the uncertainty expressed earlier, suggests that isoagglutination is acquired by the dominant effect of iso1 mutation, but can be reinstated only during zygote maturation and meiotic germination. Under this hypothesis, MID-dependent suppression of plus-related genes represents an epigenetic state determined well ahead of gametogenesis. This requirement, in turn, suggests an evolutionary scenario from homothallic to heterothallic sex-determination wherein MID acquires plus suppression functions on top of its ancestral function of minus activation. Evolution of MID functions from its ancestral role has been also noted in a sexually dimorphic oogamous species, Volvox carteri (Geng et al. 2014, 2018).
The parent diploids had been generated by double complementation of ac17 (on ch3:6393k) and nit2 (on ch3:4693k) alleles, and we found expected segregation of ac17 and nit2 phenotypes among the progeny, confirming successful meiotic germination of the tetraploid zygotes. Backcrosses to wild type mt+/mt− diploids allowed recovery of mt+/mt+ and mt−/mt− strains with or without the iso1 mutation, as confirmed by PCR-genotyping.
Our successful tetraploid genetics with iso1 diploids suggests that it may be possible to perform octaploid genetics, and so forth, testing the upper limit of genomic content in C. reinhardtii. As a first step, we mated AC17 nit2 and ac17 NIT2 homozygous diploids collected from backcross progeny. Using the same double complementation scheme used for generating diploids, we were able to collect putative tetraploids, T1 and T2 (tetraploid 1 and 2), carrying mt+/mt+/mt−/mt−, that grow normally and differentiate into minus gametes in N-free conditions. To ascertain ploidy levels, we used DNA-specific quantitation (see Materials and Methods), and compared the DNA content of T1 and T2 cells with two haploids (CC-125 and CC-124) and the parental diploids (D1, D2). The result indicated tetraploidy of T1 and T2 (Table 3).
Ploidy levels are reported to influence cell size in many organisms (Matsunaga et al. 2013). Quantitative analyses revealed that haploid cells have a median volume of 245.1 μm3, and that polyploid cells were indeed at least twice as large as haploid cells, but we did not see significant differences between D2 (median 818.1 μm3) and T1 (median 610.9 μm3) (Fig. 2). T1 cells showed larger variation in size including some extremely large cells. Our results indicate positive association of genome size and cell size in C. reinhardtii.
Our lab will provide the heterozygous iso1 diploid strains capable of selfing, which carry a pARG7 insertion at ISO1 locus that allows PCR-based genotyping and can produce wild type (ISO1/ISO1) progeny, to investigators interested in pursuing this approach (Table 4).
Meiosis induction via homeoprotein heterodimerizationStudies from the Snell laboratory (Zhao et al. 2001) established that the GSP1 gene encoding the GSP1 homeoprotein is expressed exclusively in plus gametes, and that when minus strains are transformed with GSP1 under a constitutive promoter (T-GSP1) and are then nitrogen-starved to induce gametogenesis, they express genes normally transcribed only in the diploid zygote, the result being that they produce zygotic pellicle. Our previous study (Lee et al. 2008) showed that this outcome results from the heterodimerization of the introduced GSP1 with a second homeoprotein, GSM1, that is produced exclusively by minus gametes: when GSM1/GSP1 heterodimers form, they are capable of entering the nucleus and activating transcription of the zygote program. Hence when plus strains are transformed with T-GSM1 and then nitrogen-starved, their endogenously produced GSP1 interacts with the transgene-introduced GSM1, and zygotic gene expression and wall formation proceed in haploid cells.
Extending this approach to the diploid cells generated using the iso1 protocol, we found (Lee et al. 2008) that when mt+/mt+ diploids carrying T-GSM1 or mt+/mt− diploids carrying T-GSP1 are nitrogen-starved, they not only produce thick chloroform-resistant zygotic walls but also proceed to undergo meiosis to generate haploid progeny. For reasons not yet understood, the germination process is delayed: whereas “regular” zygotes usually require ~5 days of dark incubation followed by transfer to fresh plates in the light to stimulate the onset of meiosis (Harris 1989), the transgenic diploids require ~30 days of darkness prior to transfer and light. Germination is then good to excellent (59–98%), and there occurs normal segregation of alleles and recombination of linked markers (Lee et al. 2008).
Use of T-GSP1 or T-GSM1 diploidsTransgenic introduction of Gsp1 or Gsm1 can be useful for large-scale genetic analysis. First, it allows a convenient diploid → haploid conversion (Lee et al. 2008) without going through complicated triploid matings, and second, it offers an alternative storage solution for mutant libraries.
Long-term storage of a large-scale mutant library is a significant undertaking. Current solutions include robotics-assisted periodic transfers to new cultures and freezing stocks in liquid nitrogen (Gonzalez-Ballester et al. 2011, Dent et al. 2015, Li et al. 2016), which are either not reliable or require high maintenance. Several investigators have therefore tried to subject diploid zygotes to long-term storage, either in desiccated or frozen states. As reviewed in Harris (1989), this has met with some success. However, this option has major drawbacks: the stored zygotes must be capable of undergoing a successful meiosis, and even fresh zygotes are sometimes known to be uncooperative in this regard; moreover, the meiotic progeny are recombinant so that one’s mutation or transgene of interest is now in a different genetic background from the original strain, which may become significant impediment especially when analyzing quantitative traits sensitive to allelic variations (Flint and Mackay 2009).
When haploid mt+ or mt− cells are starved for nitrogen, they form zygote-specific cell walls. When subjected to standard lab procedures for zygote maturation and germination, they digest their surrounding walls and proceed to form haploid colonies via mitosis. We have ascertained that these “haploid zygotes” survive exposure to chloroform vapors, indicating that their walls are similar if not identical to the walls of diploid zygotes. They also survive being mixed with adsorbent clay particles and dried for a week, another hallmark of diploid zygotes (Harris 1989); vegetative controls, by contrast, are uniformly killed by this drying procedure (Fig. 3). After 100 days in the desiccated condition, a subset of the dried haploid zygotes survived, suggesting the possibility of long-term storage as desiccated pellets. If the method can be verified for consistency and applicability to multiple genetic backgrounds, it should be possible to store haploid cell lines without the vagaries of maintaining them in stock tubes, a long-term goal of the Chlamydomonas community.
Available T-GSP1 or T-GSM1 diploid strains from our lab are found in Table 4. We can also provide the GSP1 and GSM1 transgenes to investigators wishing to construct their own lines.
Implications for the future of Chlamydomonas geneticsSelf-fertile haploid strainsMost model organisms possess great advantages if their reference strain or type has the capacity for selfing. Species that undergo selfing possess highly homogeneous genetic makeups that are propagated indefinitely without concern about loss, spontaneous mutations, or unknown genetic variation during backcrossing. Saccharomyces cerevisiae and Arabidopsis thaliana are two important examples.
So far, we have found three isoagglutination mutations, iso1, iso2, and iso3 (J.–H. Lee unpublished), all of which show isoagglutination only in a mt− background, and all of which fail to form zygotes since their “pseudo-plus” gametes lack the FUS1 gene. Another option would be to use a plus strain harboring transgenic MID, whose gametes also show mixed plus and minus phenotypes (Ferris and Goodenough 1997). It remains to be tested whether self-mating of mt+ T-MID cells successfully results in zygote formation and whether the selfing zygotes execute meiotic germination as efficiently as heterogametic zygotes. Homothallic species in Chlamydomonas genus have been reported and studied for its sexual and zygotic development (Lewin 1951, VanWinkle-Swift and Bauer 1982). A truly self-fertile strain in the primary model species, C. reinhardtii, will not only allow the study of quantitative traits but also facilitate large-scale genomics approach to another level. For example, genome-wide genetic interaction studies or double / triple-knockouts to study redundant genes can be programmed and executed from a gene-indexed mutant library established in a self-fertile strain by the help of robotics, since all the mutants in the library can mate with each other.
Study of essential genesRecently, several groups of investigators successfully applied CRISPR/Cas9-technology to C. reinhardtii, targeting specific loci for deletion or replacement (Baek et al. 2016, Shin et al. 2016, Greiner et al. 2017). Combining a sequence-specific knockout tool such as CRISPR/Cas9 and genetically amenable diploid strains will enable the systematic study of essential genes in C. reinhardtii.
CONCLUSIONIn this study, new techniques are described for generating and performing genetic analysis of diploid strains of Chlamydomonas reinhardtii. These protocols and available diploid strains for tetraploid genetics will facilitate the isolation, characterization, mapping, and maintenance of recessive lethal mutations. A protocol for generating haploid zygotes is also described that may abet long-term storage of mutant strains.
ACKNOWLEDGEMENTSWe thank Susan Dutcher for providing ac17 NIT2 strains for generating vegetative diploids. This work was supported by Discovery Grant 418471-12 from the Natural Sciences and Engineering Research Council (NSERC) (to J.-H.L.), by the Korea Carbon Concentration and Sequestration Research and Development Center (KCRC), Korean Ministry of Science, grant no. 2016M1A8A1925345 (to J.-H.L.).
Fig. 1The genetics workflow of Chlamydomonas reinhardtii discussed in this study. (A) Conventional diploid genetics utilizes haploid plus and minus strains that mate and differentiate into diploid zygotes, which can germinate to produce recombinant haploid progeny following meiosis. (B & C) Two methods of diploid generation for testing allelic interactions. (B) Conventional mating may produce mitotic diploids (mt+/mt−) at low frequency (<1%), which can be collected by dual selection, exhibiting minus sexual phenotype. (C) Polyethylene glycol (PEG)-mediated protoplast fusion combines any combination of haploid strains. The resulting hybrid strains may exhibit aneuploidy and genomic instability. (D) Tetraploid genetics utilizes self-mating heterozygous diploids (ISO1/iso1 mt+/mt−) carrying the dominant iso1 mutation, which yield tetraploid zygotes that germinate to produce recombinant diploid progeny of any mating type combination via meiosis. (E) Haploidization of diploids can be induced by T-GSM1 or T-GSP1, activating zygote development entailing meiosis in the absence of mating. (F) Pseudozygote-spores can be produced by T-GSM1 or T-GSP1 without mating, offering a long-term storage solution for C. reinhardtii strains. 
Fig. 2Polyploidy cells are bigger than haploid cells. Logarithmic phase cultures of CC-124 (1n, n = 139), D2 (2n, n = 139), T1 (4n, n = 98) were collected for image analysis. Gray box shows upper and lower quantiles (75% and 25% of the given data) and median values with a thick horizontal line. Notches of the box indicate confidence intervals calculated by the median ± [1.58 × interquartile range / sqrt(n)]. Individual values were shown with jittering dots. Vertical lines indicate ±1.5 standard deviation. 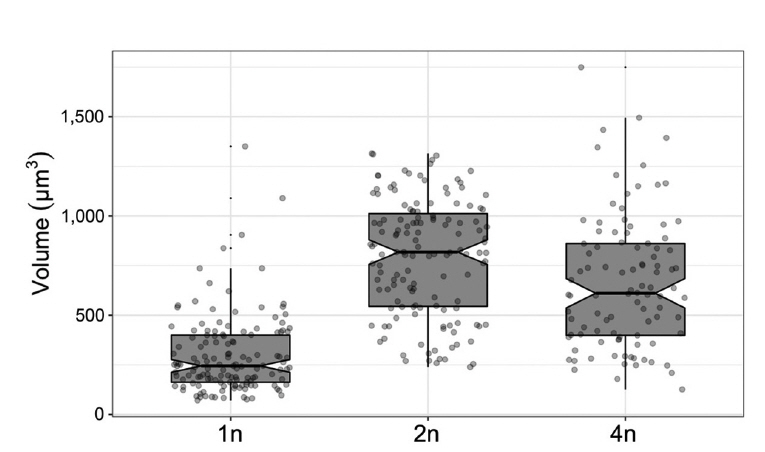
Fig. 3Desiccated haploid zygotes survived after 100 days in storage. WT strains: +, CC-125; −, CC-124. T-GSM1 strains: +, a and b; +/+, M1-homo. T-GSP1 strains: −, a and c; +/−, P1. M+P strains harboring both T-GSM1 and T-GSP1: +, MP-plus; −, MP-minus. Haploid and diploid transgenics lines harboring T-GSM1 and / or T-GSP1 are described in Lee et al. (2008). 
Table 1List of primers for genotyping Table 2Gametic characteristics of diploids generated from haploid iso1 strains
Table 3Confirmation of ploidy in newly generated polyploidy lines in this study
REFERENCESBaek, K., Kim, DH., Jeong, J., Sim, SJ., Melis, A., Kim, J-S., Jin, E. & Bae, S. 2016. DNA-free two-gene knockout in Chlamydomonas reinhardtii via CRISPR-Cas9 ribonucleoproteins. Sci Rep. 6:30620 pp.
Bellafiore, S., Ferris, P., Naver, H., Göhre, V. & Rochaix, JD. 2002. Loss of Albino3 leads to the specific depletion of the light-harvesting system. Plant Cell. 14:2303–2314.
Blaby, IK., Blaby-Haas, CE., Tourasse, N., Hom, EF., Lopez, D., Aksoy, M., Grossman, A., Umen, J., Dutcher, S., Porter, M., King, S., Witman, GB., Stanke, M., Harris, EH., Goodstein, D., Grimwood, J., Schmutz, J., Vallon, O., Merchant, SS. & Prochnik, S. 2014. The Chlamydomonas genome project: a decade on. Trends Plant Sci. 19:672–680.
Campbell, AM., Rayala, HJ. & Goodenough, UW. 1995. The iso1 gene of Chlamydomonas is involved in sex determination. Mol Biol Cell. 6:87–95.
Dent, RM., Sharifi, MN., Malnoë, A., Haglund, C., Calderon, RH., Wakao, S. & Niyogi, KK. 2015. Large-scale insertional mutagenesis of Chlamydomonas supports phylogenomic functional prediction of photosynthetic genes and analysis of classical acetate-requiring mutants. Plant J. 82:337–351.
Dutcher, SK. 1988. Nuclear fusion-defective phenocopies in Chlamydomonas reinhardtii: mating-type functions for meiosis can act through the cytoplasm. Proc Natl Acad Sci U S A. 85:3946–3950.
Ferris, PJ. & Goodenough, UW. 1997. Mating type in Chlamydomonas is specified by mid, the minus-dominance gene. Genetics. 146:859–869.
Ferris, PJ., Woessner, JP. & Goodenough, UW. 1996. A sex recognition glycoproteins is encoded by the plus mating-type gene fus1 of Chlamydomonas reinhardtii
. Mol Biol Cell. 7:1235–1248.
Flint, J. & Mackay, TF. 2009. Genetic architecture of quantitative traits in mice, flies, and humans. Genome Res. 19:723–733.
Gallaher, SD., Fitz-Gibbon, ST., Glaesener, AG., Pellegrini, M. & Merchant, SS. 2015.
Chlamydomonas genome resource for laboratory strains reveals a mosaic of sequence variation, identifies true strain histories, and enables strain-specific studies. Plant Cell. 27:2335–2352.
Galloway, RE. & Goodenough, UW. 1985. Generic analysis of mating locus linked mutations in Chlamydomonas reinhardtii
. Genetics. 111:447–461.
Galloway, RE. & Holden, LR. 1984. Transmission and recombination of chloroplast genes in asexual crosses of Chlamydomonas reinhardii. I. Flagellar agglutination prior to fusion does not promote uniparental inheritance or affect recombination frequencies. Curr Genet. 8:399–405.
Geng, S., De Hoff, P. & Umen, JG. 2014. Evolution of sexes from an ancestral mating-type specification pathway. PLoS Biol. 12:e1001904 pp.
Geng, S., Miyagi, A. & Umen, JG. 2018. Evolutionary divergence of the sex-determining gene MID uncoupled from the transition to anisogamy in volvocine algae. Development. 145:dev162537 pp.
Gonzalez-Ballester, D., Pootakham, W., Mus, F., Yang, W., Catalanotti, C., Magneschi, L., de Montaigu, A., Higuera, JJ., Prior, M., Galván, A., Fernandez, E. & Grossman, AR. 2011. Reverse genetics in Chlamydomonas: a platform for isolating insertional mutants. Plant Methods. 7:24 pp.
Greiner, A., Kelterborn, S., Evers, H., Kreimer, G., Sizova, I. & Hegemann, P. 2017. Targeting of photoreceptor genes in Chlamydomonas reinhardtii via zinc-finger nucleases and CRISPR/Cas9. Plant Cell. 29:2498–2518.
Harris, EH. 1989. The Chlamydomonas sourcebook: a comprehensive guide to biology and laboratory use. Academic Press, San Diego, CA, 780 pp.
Lee, JH., Lin, H., Joo, S. & Goodenough, U. 2008. Early sexual origins of homeoprotein heterodimerization and evolution of the plant KNOX/BELL family. Cell. 133:829–840.
Li, X., Zhang, R., Patena, W., Gang, SS., Blum, SR., Ivanova, N., Yue, R., Robertson, JM., Lefebvre, PA., Fitz-Gibbon, ST., Grossman, AR. & Jonikas, MC. 2016. An indexed, mapped mutant library enables reverse genetics studies of biological processes in Chlamydomonas reinhardtii
. Plant Cell. 28:367–387.
Lin, H. 2006. Characterization of two minus-specific genes, MID and MTD1, and a sex-limited mutant, ISO1, involved in Chlamydomonas gametogenesis. PhD dissertation. Washington University, St. Louis, MO, USA, 190 pp.
Lin, H., Cliften, PF. & Dutcher, SK. 2018. MAPINS, a highly efficient detection method that identifies insertional mutations and complex DNA rearrangements. Plant Physiol. 178:1436–1447.
Lin, H. & Goodenough, UW. 2007. Gametogenesis in the Chlamydomonas reinhardtii minus mating type is controlled by two genes, MID and MTD1
. Genetics. 176:913–925.
Matagne, RF., Deltour, R. & Ledoux, L. 1979. Somatic fusion between cell wall mutants of Chlamydomonas reinhardi
. Nature. 278:344–346.
Matsunaga, S., Katagiri, Y., Nagashima, Y., Sugiyama, T., Hasegawa, J., Hayashi, K. & Sakamoto, T. 2013. New insights into the dynamics of plant cell nuclei and chromosomes. Int Rev Cell Mol Biol. 305:253–301.
Merchant, SS., Prochnik, SE., Vallon, O., Harris, EH., Karpowicz, SJ., Witman, GB., Terry, A., Salamov, A., Fritz-Laylin, LK., Maréchal-Drouard, L., Marshall, WF., Qu, LH., Nelson, DR., Sanderfoot, AA., Spalding, MH., Kapitonov, VV., Ren, Q., Ferris, P., Lindquist, E., Shapiro, H., Lucas, SM., Grimwood, J., Schmutz, J., Cardol, P., Cerutti, H., Chanfreau, G., Chen, CL., Cognat, V., Croft, MT., Dent, R., Dutcher, S., Fernández, E., Fukuzawa, H., González-Ballester, D., González-Halphen, D., Hallmann, A., Hanikenne, M., Hippler, M., Inwood, W., Jabbari, K., Kalanon, M., Kuras, R., Lefebvre, PA., Lemaire, SD., Lobanov, AV., Lohr, M., Manuell, A., Meier, I., Mets, L., Mittag, M., Mittelmeier, T., Moroney, JV., Moseley, J., Napoli, C., Nedelcu, AM., Niyogi, K., Novoselov, SV., Paulsen, IT., Pazour, G., Purton, S., Ral, JP., Riaño-Pachón, DM., Riekhof, W., Rymarquis, L., Schroda, M., Stern, D., Umen, J., Willows, R., Wilson, N., Zimmer, SL., Allmer, J., Balk, J., Bisova, K., Chen, CJ., Elias, M., Gendler, K., Hauser, C., Lamb, MR., Ledford, H., Long, JC., Minagawa, J., Page, MD., Pan, J., Pootakham, W., Roje, S., Rose, A., Stahlberg, E., Terauchi, AM., Yang, P., Ball, S., Bowler, C., Dieckmann, CL., Gladyshev, VN., Green, P., Jorgensen, R., Mayfield, S., Mueller-Roeber, B., Rajamani, S., Sayre, RT., Brokstein, P., Dubchak, I., Goodstein, D., Hornick, L., Huang, YW., Jhaveri, J., Luo, Y., Martínez, D., Ngau, WC., Otillar, B., Poliakov, A., Porter, A., Szajkowski, L., Werner, G., Zhou, K., Grigoriev, IV., Rokhsar, DS. & Grossman, AR. 2007. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 318:245–250.
Molnar, M. & Sipiczki, M. 1993. Polyploidy in the haplontic yeast Schizosaccharomyces pombe: construction and analysis of strains. Curr Genet. 24:45–52.
Palombella, AL. & Dutcher, SK. 1998. Identification of the gene encoding the tryptophan synthase β-subunit from Chlamydomonas reinhardtii
. Plant Physiol. 117:455–464.
Pan, X., Yuan, DS., Xiang, D., Wang, X., Sookhai-Mahadeo, S., Bader, JS., Hieter, P., Spencer, F. & Boeke, JD. 2004. A robust toolkit for functional profiling of the yeast genome. Mol Cell. 16:487–496.
Romero-Campero, FJ., Perez-Hurtado, I., Lucas-Reina, E., Romero, JM. & Valverde, F. 2016. ChlamyNET: a Chlamydomonas gene co-expression network reveals global properties of the transcriptome and the early setup of key co-expression patterns in the green lineage. BMC Genomics. 17:227 pp.
Shin, S-E., Lim, J-M., Koh, HG., Kim, EK., Kang, NK., Jeon, S., Kwon, S., Shin, W-S., Lee, B., Hwangbo, K., Kim, J., Ye, SH., Yun, J-Y., Seo, H., Oh, H-M., Kim, K-J., Kim, J-S., Jeong, W-J., Chang, YK. & Jeong, B-R. 2016. CRISPR/Cas9-induced knockout and knock-in mutations in Chlamydomonas reinhardtii
. Sci Rep. 6:27810 pp.
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||