INTRODUCTION
Distribution patterns (both intra- and inter-specific) are influenced by many biotic and abiotic factors working at different time scales. Patterns of species diversity (species assemblages) are influenced by factors that work over large temporal scales, whereas factors that influence patterns within conspecific populations work on shorter time scales (Avise 2006). Broad-scale abiotic factors (e.g., oceanic currents, temperature) influence many species and lead to patterns of species assemblages that can be characterized as biogeographic regions (regions of species assemblages different from other areas). The differences between biogeographic regions are often due to major and long term differences in environments or longterm barriers to dispersal. Biogeographic classification based on species assemblages have also been applied in marine environments which is theory are contiguous.
In New Zealand several marine biogeographic classification schemes have been proposed (summarized in Shears et al. 2008), one based mostly on presence-absence of macroalgal species (Shears et al. 2008). Currently it is used, in conjunction with other data (e.g., physical and oceanographic data) to classify New Zealand marine environment into fourteen biogeographic regions, for conservation and protection (Department of Conservation, Ministry of Fisheries 2008, 2011). These schemes assumes that physical habitats and ecosystems, will contain different biological communities due to a combination of broad-scale factors, such as water temperature, oceanography, current dynamics and habitat types.
A review of molecular studies on the phylogeography of coastal marine invertebrates and plants in New Zealand, revealed a multitude of phylogeographic breaks (Ross et al. 2009 and references herein), none of which was fully congruent with all the bioregions used by governmental agencies (Department of Conservation, Ministry of Fisheries 2011). However, two genetic breaks are of special interest as they have been found frequently. One is the biogeographic break at East Cape, on the northeast of the North Island, where oceanic eddies are believed to obstruct connectivity between adjacent populations (Stevens and Hogg 2004, Shears et al. 2008). The other is the phylogeographic division between the top of the South Island of New Zealand, at Cape Campbell on the northeast coast of the South Island, and regions south of this. Upwelling south of Cape Campbell was initially postulated as the driving force (Waters and Roy 2004b, Ayers and Waters 2005), but a recalibration of the divergence time in limpets showed that the genetic breaks were older than the upwelling zone (Goldstien et al. 2006). Changes in the hydrography of the Cook Strait region during the late Pleistocene as a consequence of fluctuating sea levels following the cycles of glaciation has been suggested as alternative causes (Goldstien et al. 2006), although continued comparative data is needed.
The genus Lessonia Bory (Laminariales, Phaeophyceae) is a prominent kelp in the southern hemisphere. Recent molecular studies have shown that the Australasian Lessonia species (L. adamsiae Hay, L. brevifolia J. Agardh, L. corrugata Lucas, L. tholiformis Hay, and L. variegata J. Agardh) are a monophyletic group and have diverged recently and rapidly, about 3.5-3 million years ago (Mya) (Martin and Zuccarello 2012). L. variegata, earlier believed to be a single species occupying the coasts of the three main islands of New Zealand (North Island, South Island, and Stewart Island) consists of a non-monophyletic species complex of four different species (Martin and Zuccarello 2012). In biogeographic surveys, L. variegata was entered as a single widely dispersed species (Shears et al. 2008).
Barriers to dispersal, lead both to population differentiation and if maintained could lead to speciation of neighboring populations. These barriers are especially effective on algae that do not have good dispersal. Lessonia is one such alga as it lacks buoyant structures (i.e., pneumatocysts), and at least L. variegata has massive holdfasts (Nelson 2013). In Chile, Lessonia nigrescens Bory was considered to be a wide ranging species distributed from central Chile to Tierra del Fuego. It is in fact composed of three lineages, recognized as cryptic species initially, each with more limited ranges and non-overlapping distributions (Tellier et al. 2009, 2011a, 2011b). These cryptic species later were given formal species names distinct from true L. nigrescens (L. spicata Suhr) Santelices and L. berteroana Montagne (González et al. 2012). The break in species distribution between these species of Lessonia in Chile appears to coincide with major biogeographic breaks at about 42-43° S and 30-33° S (Tellier et al. 2009, Guillemin et al. 2016).
The aim of this research is to determine the distribution in Lessonia species with respect to proposed bioregions and to see if these distribution patterns reflect established bioregions. Thus, we focus on the distribution of the L. variegata cryptic species around the main islands of New Zealand and discuss possible phylogeographic scenarios.
MATERIALS AND METHODS
Sampling
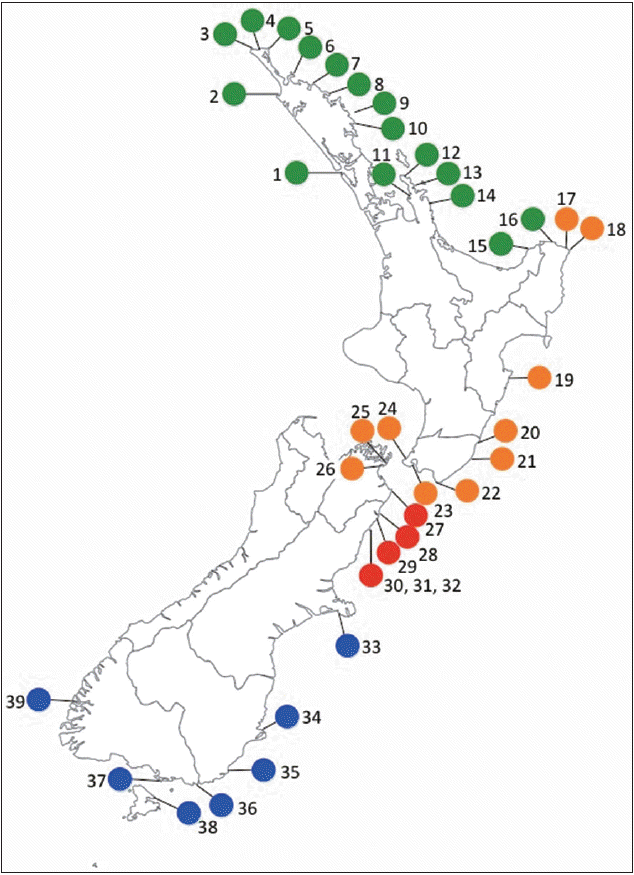
A total of 485 individuals of Lessonia were collected in the intertidal or subtidal while free diving at 59 sites in the main islands of New Zealand, plus the Chatham Islands and adjacent subantarctic islands (Snares Island, Auckland Islands, Campbell Island, and Bounty Islands) (389 individuals of L. variegata, 86 samples of the three other Lessonia species in New Zealand waters [L. adamsiae, L. brevifolia, and L. tholiformis], plus 10 samples of L. corrugata from Tasmania, Australia, part of the monophyletic western Pacific Ocean Lessonia species) (Table 1) (Martin and Zuccarello 2012). Collection sites in New Zealand’s main islands have been selected to span most phylogeographic regions as identified by Shears et al. (2008) (Fig. 1). Tissue (2-3 cm2) was collected near the transition zone at the base of the blade where the blade is youngest and free of epiphytes, patted dry and sealed in bags with silica gel.
DNA extraction was performed from approximately 0.5 cm2 of dried tissue using a modified cetyltrimethylammonium bromide (CTAB) protocol (Zuccarello and Lokhorst 2005). Polymerase chain reaction (PCR) amplification and PCR conditions of the mitochondrial spacer region between adenosine-tri-phosphate dehydrogenase subunit 8 and t-RNA serine (atp8-sp), which includes 40 bp of the atp8-gene and 17 bp of the downstream trnSgene, and the plastid spacer between the large and short subunit of the ribulose-1,5-bisphosphate carboxylase/ oxygenase gene (rbc-sp) were performed as described in Martin and Zuccarello (2012).
To identify the atp8-sp haplotypes in the many samples both among and within populations haplotypes were scored by single stranded conformational polymorphism (SSCP) (Zuccarello et al. 1999). The protocols for SSCP and silver staining are described in Buchanan and Zuccarello (2012) and Bassam et al. (1991), respectively. Known haplotypes were run as identifiers on each gel. Band patterns that deviate from the patterns of the known haplotypes were sequenced to recognize possible new atp8-sp haplotypes. Additionally, random identical SSCP bands of atp8-sp were sequenced as a control for SSCP accuracy.
Sequencing was done commercially (Macrogen Inc., Seoul, Korea) after the PCR products had been purified with ExoSAP-IT (USB, Cleveland, OH, USA). Atp8-sp sequences of 180 individuals were combined with SSCP patterns of 305 individuals for further analysis, and 81 individuals were processed with both methods to check SSCP reliability. Additionally 46 individuals were also sequenced with a plastid marker, rbc-sp region.
Forward and reverse sequences were assembled in the ‘STADEN package’ (Staden et al. 2001) using the implementable programs PHRED (Ewing and Green 1998), PHRAP (Green unpublished, http://bozeman.genome.washington.edu/phrap.docs/phrap.html), and GAP4 (Bonfield et al. 1995). The alignment was performed in BIOEDIT ver. 7.0.9.0 (Hall 1999) using ClustalW (Larkin et al. 2007) and was subsequently refined by eye.
TCS 1.21 (Clement et al. 2000) was used to construct a haplotype network based on atp8-sp and rbc-sp sequence data. The program Modeltest version 3.7 (Posada and Crandall 1998) was used to find the model of sequence evolution that best fit the data set by an Akaike Information Criterion (AIC) (Posada and Crandall 2001). Maximum likelihood was performed with RAxML 7.2.8 (Stamatakis 2006). The GTR + gamma model and 500 non-parametric bootstrap replicates (Felsenstein 1985) were implemented. For the atp8-sp phylogenetic reconstruction Lessonia vadosa, from Chile, was used as an outgroup, as it is sister to the west Pacific samples (Martin and Zuccarello 2012). For the rbc-sp phylogenetic reconstruction Lessonia nigresens, also from Chile, was used as an outgroup. Bayesian inference was performed with MrBayes v3.2 (Ronquist et al. 2012). Analyses consisted of two independent simultaneous runs of one cold and three incrementally heated chains, and 3 × 106 generations with sampling every 1,000 generations. The log files of the runs were checked with Tracer v1.5 (Rambaut and Drummond 2009) and a burn-in sample of 500 trees was removed from each run before calculating the majority rule consensus tree.
RESULTS
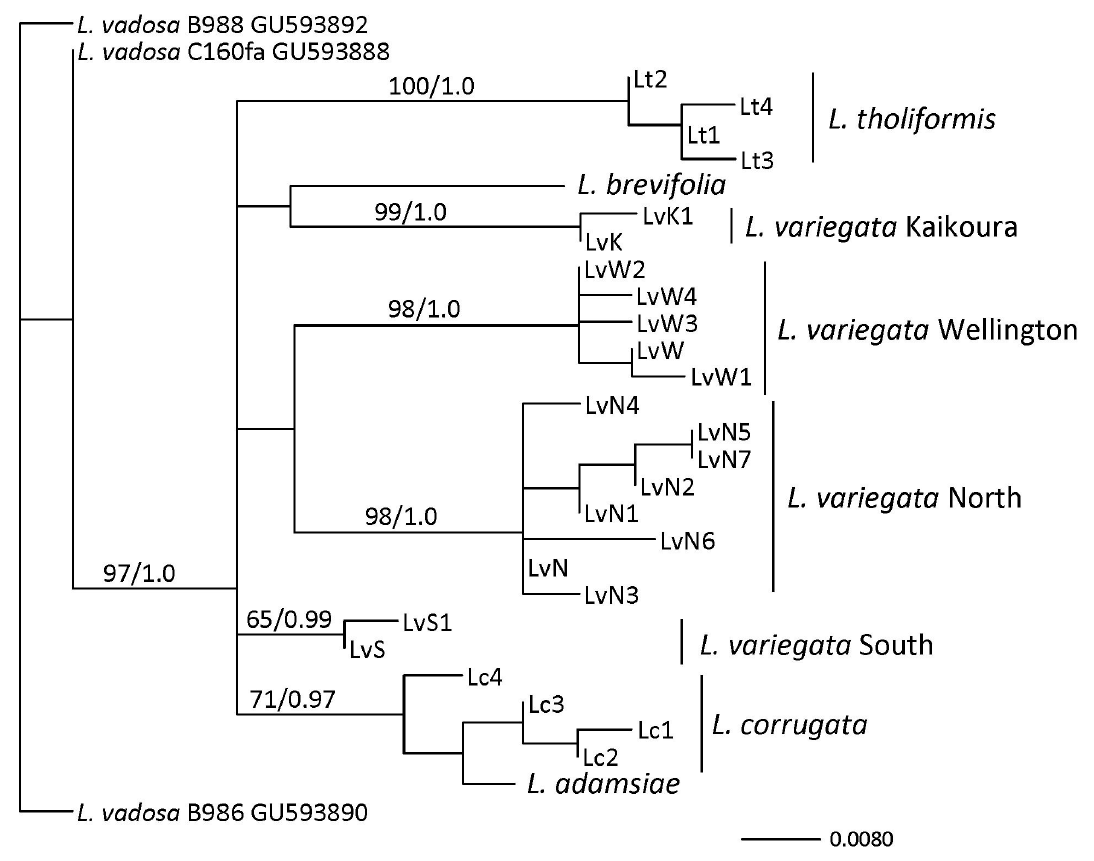
Previous studies have shown that L. variegata consist of four cryptic species (Martin and Zuccarello 2012), identified as L. variegata / N (for the northern species), L. variegata / W (for the species around Wellington), L. variegata / K (for the species around Kaikoura, South Island), and L. variegata / S (for the southern species). We will use these designations here. All SSCP banding and sequencing results were concordant.
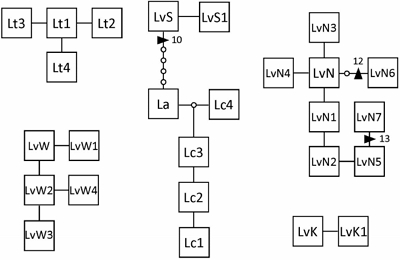
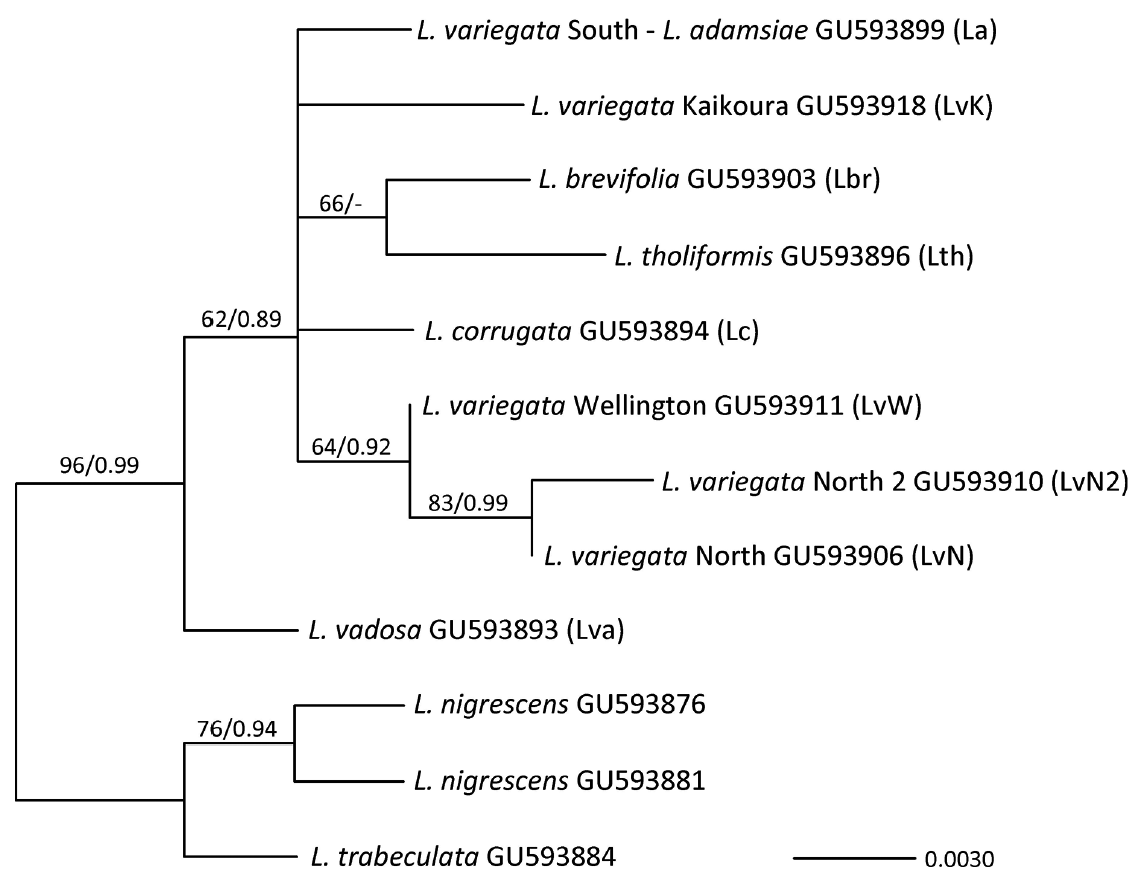
The mtDNA atp8-sp yielded an aligned dataset of 198 bp. There was no (L. adamsiae and L. brevifolia) to little (L. variegata / S and L. variegata / K; 2 haplotypes each) variation in these Lessonia species, and the highest variation was in L. variegata / N (8 haplotypes). The other species showed intermediate variation (Fig. 2) (Genbank accession Nos. KX023377-KX023403). A connected network was seen between L. variegata / S, L. adamsiae, and L. corrugata (separated by at least 5 bp substitutions and one 10 bp indel), while the relationship between L. variegata / S and L. adamsiae / L. corrugata was not supported in the phylogenetic analysis (Appendix 1). Overall the relationships between the species found in Australasian waters were not resolved with this data set but are similar to the results reported with multiple loci (Martin and Zuccarello 2012), while even with this dataset most species were monophyletic (Appendix 1). Within the eight haplotypes of L. variegata / N there were two indels (12 bp and 13 bp) (Fig. 2).
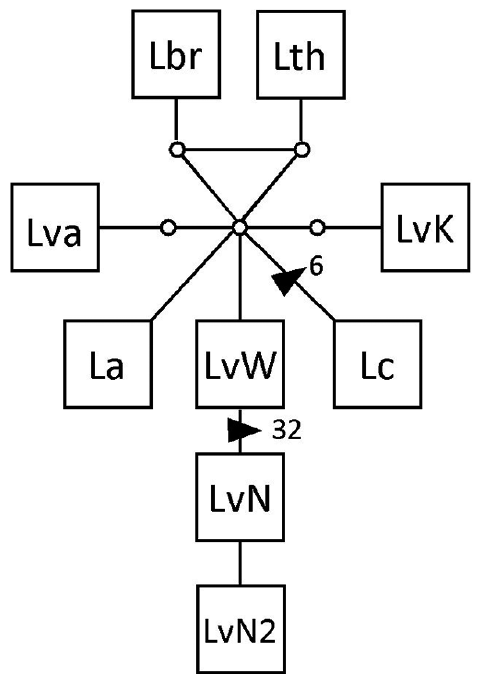
The cpDNA rbc-sp yielded a dataset of 365 bp. This marker was more conserved than the atp8-sp (Martin and Zuccarello 2012) and only showed variation within L. variegata / N (2 haplotypes). The L. variegata / N haplotypes differ by a shared 32 bp indel from the L. variegata / W haplotype (Appendix 2). L. variegata / S and L. adamsiae shared a rbc-sp haplotype otherwise all species had distinct haplotypes. The network of the rbc-sp for Australasian Lessonia showed a star-like pattern with a missing central haplotype (Appendix 2). The phylogenetic reconstruction was unresolved but showed a moderately supported monophyletic L. variegata / N and L. variegata / W (Appendix 3).
All four L. variegata cryptic species displayed a nonoverlapping distribution (i.e., endemism) around New Zealand (Fig. 1). L. variegata / N is found from Hokianga, west coast of the North Island (site 1) around North Cape (top of the North Island) to Lottin Point, East Cape (site 16). A break was found around East Cape between L. variegata / N at Lottin Point and L. variegata / W (Horoera, site 17), two sites approximately 35 km apart. L. variegata / W was found south from Horoera all along the southeast coast of the North Island to Whites Bay (Marlborough Sounds / South Island, site 26), spanning Cook Strait (between the North and South Island) and several sites in the Wellington Region (grouped as site 23) (Table 1). The break between L. variegata / W (site 26) and L. variegata / K (site 27) was approximately 50 km composed mostly of sandy shores. L. variegata / K was solely found between Cape Campbell (site 27) to Paia Point (17 km south of Kaikoura, site 32). L. variegata / S was found south from Tumbledown Bay, Banks Peninsula (site 33) to Doubtful Sound (site 39) including Stewart Island (site 38).
The other species of Lessonia were restricted to particular off-shore islands or Australia. L. adamsiae was found on the Snares Island. L. brevifolia was found on several island groups: Auckland, Campbell, Antipodes, and Bounty. L. tholiformis was collected from several sites around the Chatham Islands, and L. corrugata from seven sites around Tasmania, Australia. Only L. corrugata and L. tholiformis showed any variation in atp8-sp (4 haplotypes each), all the other species were not variable with this marker.
DISCUSSION
We report on the distribution of a supposedly widely distributed species, L. variegata, in New Zealand. This ‘species’ is composed of four distinct lineages, that can be considered cryptic species (Martin and Zuccarello 2012), that have non-overlapping distributions. This pattern of widely distributed Lessonia ‘species’ being comprised of multiple cryptic species, with mostly non-overlapping distributions has been shown in the L. nigrescens species complex in Chile (Tellier et al. 2009, 2011b, González et al. 2012). The genetic breaks or species boundaries found in the L. variegata species complex roughly correspond to breaks proposed between biogeographic regions in New Zealand (Shears et al. 2008, Department of Conservation, Ministry of Fisheries 2011). While this data set suggests that L. adamsiae and L. corrugata may be conspecific, previous data, using a multi-gene approach, suggests that they are distinct sister species (Martin and Zuccarello 2012).
L. variegata / N is found across three bioregions (Appendix 4), throughout the Northeastern bioregion, but on the western boundary it was found as far south as Hokianga (site 1) (West Coast North Island region). On the eastern side a sharp genetic break between L. variegata / N and L. variegata / W (between sites 16 and 17, less than 35 km apart) corresponds to the break between the North Eastern and East Coast North Island bioregion (Department of Conservation, Ministry of Fisheries 2011). This break, in the region of East Cape (north east North Island), has been identified in a multitude of biogeographic classification schemes (summarized in Shears et al. 2008) and phylogeographic studies (Stevens and Hogg 2004, Waters and Roy 2004b, Ayers and Waters 2005, Ross et al. 2009), and is probably the most detected biogeographic break in the marine realm around New Zealand. The break is generally attributed to the permanent and anticyclonic East Cape Eddy that captures most of the south east flowing East Auckland current and takes it off shore (Stanton et al. 1997) and is thus believed to be a significant barrier to gene flow (e.g., Stevens and Hogg 2004). Despite the retention effect of the East Cape Eddy, temperature differences between the warm subtropical East Auckland Current and the cool northward flowing Wairarapa Current, which meet at East Cape (Chiswell 2000) are also evident, these may also maintain species distribution boundaries. Further sampling in this region will identify the small-scale pattern of distribution of these two cryptic species, and if the break is similar to the mosaic distribution, plus lack of hybridization, as found in L. nigrescens species complex in Chile (Tellier et al. 2011a, 2011b).
L. variegata / W is found from East Cape (site 17) to the northern end of the South Island (site 26). This distribution crosses a biogeographic break, between the East Coast North Island and North Cook Strait bioregions (Department of Conservation, Ministry of Fisheries 2011) (Appendix 4). Several phylogeographic studies have not detected genetic breaks either along the south east coast of the North Island, nor across Cook Strait (Ayers and Waters 2005, Goldstien et al. 2006), including in the algae Bostrychia intricata (Bory) Montagne (Muangmai et al. 2015) and Carpophyllum maschalocarpum (Turner) Greville (Buchanan and Zuccarello 2012).
In concordance with Department of Conservation, Ministry of Fisheries (2011) a genetic break near Cape Campbell between L. variegata / W and L. variegata / K was found (between sites 26 and 27). A break near Cape Campbell was previously found in several phylogeographic surveys (Apte and Gardner 2002, Stevens and Hogg 2004, Waters and Roy 2004b, Ayers and Waters 2005, Goldstien et al. 2006). Whereas some suggested upwelling as a possible differentiating force (e.g., Apte and Gardner 2002, Waters and Roy 2004b, Ayers and Waters 2005) or favored currents and eddies (Stevens and Hogg 2004), others suggested that changes in the dynamic hydrography of Cook Strait triggered differentiation (Goldstien et al. 2006). A molecular dating analysis showed that the postglacial upwelling south of Cape Campbell originated long after the genetic differentiation in Patiriella regularis Verrill, and therefore did not support upwelling as a cause (Goldstien et al. 2006). Upwelling can also not explain the break in the two Lessonia species as L. variegata / K is found across the area of upwelling, which is usually assumed to be just south of Cape Campbell. Whether currents and eddies are a likely barrier is not known. The break between L. variegata / W and L. variegata / K also corresponds with marshland from two successive river deltas, along a distance of more then 40 km, this probably makes for unsuitable habitat in this area.
L. variegata / K is solely found in the northeast of the South Island from Cape Campbell (site 27) to Paia Point (site 32), the East Coast South Island bioregion (Appendix 4). The genetic break between the southernmost L. variegata / K samples and the northernmost L. variegata / S populations (site 33) is also an area dominated by sandy shores. In both this and the previous case, short (less than 40 km) expanses of unsuitable habitat appear to be associated with changes in species distribution. These short distance breaks are different from what is seen from algae with floating structures (Buchanan and Zuccarello 2012) and consistent with lack of dispersal over short distance in Lessonia (Tellier et al. 2011a).
L. variegata / S is distributed throughout the southern South Island, over to Stewart Island to the west coast (Fiordland bioregion). No Lessonia was found along the sandy areas that characterize the majority of the southeast coast of the South Island except for the population on Banks Peninsula (site 33). As this population is more than 200 km north of its nearest neighbor (site 34), it is possibly better described as a satellite population. The sandy coast line in this area may be a strong barrier to gene flow for many benthic organisms that need hard substrate.
Our results show high phylogeographic structure and local endemism in Lessonia. All four species within the L. variegata species complex, are confined to certain areas with no overlap in their distribution. How did these species speciate? Multiple-gene phylogenies and molecular clock analysis suggests that there was a rapid radiation in the Australasian Lessonia that took place 3.5-3 Mya, over 3-5 k years (Martin and Zuccarello 2012). A sudden radiation is often indicative of dramatic changes in the history of a species (e.g., the time of colonisation into a new area via long distance dispersal) (Waters and Roy 2004a). At the time of radiation in Lessonia (mid to late Pliocene), the central and southern part of the North Island was mainly submerged (Manawatu Strait) and a ridge separated northeast North Island from what would become the southeastern North Island (Trewick and Bland 2012). L. variegata / W possibly has its origin on this ridge before it later became connected to the northern North Island about 1 Mya (Bunce et al. 2009). The East Cape Eddy could then have maintained separation between the two North Island species even after the closure of the Manawatu Strait.
Apart from L. variegata / W on the northeast coast of the South Island, two additional Lessonia species are found on the South Island. One, L. variegata / K is endemic to the northeast from Cape Campbell to Paia Point. Vicariance (i.e., the formation of Cook Strait) was unlikely to be the cause of speciation between the ancestor of L. variegata / W and L. variegata / K, especially as the timing of their radiation (Martin and Zuccarello 2012) predates the opening of Cook Strait (Fleming 1979).
On its southern limit L. variegata / K is separated from L. variegata / S by a >350 km gap of quaternary sediment. This gap formed by glacial outwash with the retreat of the first ice age of the Pleistocene and has persisted during glacial and interglacial period ever since (Fleming 1979). Sandy beaches and soft sandstones are unsuitable habitat for Lessonia (personal observation) and no Lessonia was found in these intervening areas. In L. nigrescens, it was suggested that the normal dispersal potential in Lessonia is low (e.g., L. nigrescens) (Tellier et al. 2011b). Thus this unsuitable habitat is probably large enough to prevent connectivity and enabled divergent evolution between these two South Island species.
In conclusion, our data suggests that L. variegata species have a distribution that mostly follows biogeographic schemes proposed for New Zealand. These patterns are possibly due to both ancient geological processes (island splitting, sea level changes) and more contemporary oceanographic processes (currents and eddies). Our results and other research also suggest that Lessonia is a poor disperser (Tellier et al. 2011b), in contrast to other kelps like Macrocystis (Macaya and Zuccarello 2010) and Durvillaea (Fraser et al. 2009), and therefore has had more opportunity to speciate. The dispersal capacity of Lessonia in New Zealand needs further investigation.