INTRODUCTION
The foliose Bangiaceae (Engler 1892) is a diverse, cosmopolitan, and distinctive family of morphologically simple red algae that represent an ancient lineage (Butterfield 2000), and economically important seaweeds used as resources in aquaculture for the production of ‘gim’ or ‘nori’ in Korea and Japan, respectively (Sutherland et al. 2011, Kim et al. 2017). Identification and systematics within the family, however, have created notorious challenges because of the simple gross morphology with morphological plasticity between individulas. Traditionally, two genera have been recognized in this family, unbranched filamentous thalli assigned to the genus Bangia Lyngbye (1819) and those that are foliose assigned to the genus Porphyra C. Agardh (1824), before the revision based on a two-gene molecular phylogeny (Sutherland et al. 2011). Currently, Porphyra sensu lato is subdivided into nine genera: Boreophyllum, Clymene, Fuscifolium, Lysithea, Miuraea, Porphyra, Pyropia, Wildemania, and Neothemis (Sutherland et al. 2011, Sánchez et al. 2014, 2015). Revision at the generic level has allowed for a broad understanding of relationships within the family, and the next logical step is to examine species-level taxonomy in detail (Kucera and Saunders 2012, Lindstrom et al. 2015a, 2015b, Guillemin et al. 2016).
Species delineation within foliose Bangiaceae based on morphological analysis alone has proven to be mostly unreliable as it is in many cases for other algal groups (Kucera and Saunders 2012, Sánchez et al. 2014, Verbruggen 2014, Guillemin et al. 2016): combined morphology and molecular studies are needed. For example, approximately 46% of the foliose specimens (58 out of 126) used in the two gene phylogeny study of Sutherland et al. (2011) were undescribed or with uncertain species identification. Therefore, following extensive studies have focused on the discovery of cryptic new species and correctly identifying local specimens with the application of molecular techniques (e.g., Mateo-Cid et al. 2012, Mols-Mortensen et al. 2012, Nelson 2013, Nelson and D’Archino 2014, Ramírez et al. 2014, Sánchez et al. 2014, Harden et al. 2015, Lindstrom et al. 2015a, 2015b). As a result, 18 species of Pyropia have been newly described among the currently accepted 70 species (excluding 4 infraspecific) names after revision by Sutherland et al. (2011) (see Guiry and Guiry 2017).
Studies have employed a variety of molecular markers for the taxa of Bangiaceae including: SSU-rDNA (e.g., Stiller and Waaland 1993, Kunimoto et al. 1999, Teasdale et al. 2009), rbcL (e.g., Lindstrom and Fredericq 2003, Lindstrom 2008, Kucera and Saunders 2012, Mols-Mortensen et al. 2012, 2014, Xie et al. 2015), COI-5P (e.g., Kucera and Saunders 2012, Milstein et al. 2012, Xie et al. 2015, Guillemin et al. 2016, Dumilag and Monotilla 2017), and SSU-rDNA + rbcL (e.g., Broom et al. 2010, Sutherland et al. 2011, Mateo-Cid et al. 2012, Sánchez et al. 2014, López-Vivas et al. 2015, Milstein et al. 2015). It was estimated that the mutation rates in mitochondrial DNA causing silent-site divergence between the closely related species Porphyra purpurea and P. umbilicalis were three times faster than that of their plastid DNA and five times that of their nuclear DNA (Smith et al. 2012). Thus, the mitochondrial COI-5P marker is more effective for distinguishing the closely related species of foliose Bangiaceae. The chloroplast rbcL gene, however, has been used more widely to assess species diversity of Bangiaceae, and there are more available reference sequences that have been registered in GenBank at present. Therefore, we selected the rbcL gene to assess the Korean representatives in this study.
The foliose Bangiaceae of Korean representatives were revised based on characteristics of morphology and distribution (Hwang and Lee 2001, Kim and Kim 2011), and up 20 foliose species were listed, including two uncertain records and one forma (Kim et al. 2013). Although Korean specimens of six species of Pyropia and one unidentified Wildemania were included in the two-gene molecular phylogeny (Sutherland et al. 2011), most have yet to by their distributional range and morphology. In our preparatory molecular work of Korean foliose Bangiaceae after a monographic study (Kim and Kim 2011), several unreported or cryptic species were found. Most of them are restricted to the east coast of Korea and northern Japan based on rbcL sequences data (unpublished). In this research, we present analyses of four targeted native species along the coast of Korea, P. kinositae (Yamada & Tak. Tanaka) N. Kikuchi, M. Miyata, M. S. Hwang & H. G. Choi, P. onoi (Ueda) N. Kikuchi & M. Miyata, P. moriensis (Ohmi) N. Kikuchi & M. Miyata, and P. retorta sp. nov., and we discuss the distributional characteristics as a possible result of Quaternary climatic oscillations of the East Sea (= Sea of Japan).
MATERALS AND METHODS
Collections
Specimens were collected as gametophytes on the coast of South Korea from 2004 to 2013 (Table. 1). The collected fresh field material was transferred to the laboratory in a cooler, and several distinctive types were sorted to be pressed as voucher specimens. A small portion (ca. 1.5 × 1.5 cm) near the center of a voucher specimen was removed for DNA analysis. The small portion was cleaned by removing attached diatoms or small tiny filamentous algae using a small brush or with fine forceps under a dissecting microscope; it was then rinsed several times with fresh distilled water. The clean fragments of each voucher were dried at 50°C and stored in silica gel to preserve DNA. All vouchers used in this study were deposited at the herbarium of the Gangneung-Wonju National University.
Morphology
Morphological and anatomical observations were conducted with sexually mature fresh thalli or on partially rehydrated fragments of the vouchers using 4% formalin-seawater. Transverse sections were made using a CM1850 cryostat (Leica Microsystems, Heidelberg, Germany), but cutting sections by hand with a stainless-steel razor blade was more practical in many cases. Photomicrographs were taken with an Olympus DP70 digital camera coupled to an Olympus BX50 light microscope (Olympus, Tokyo, Japan).
DNA extraction and polymerase chain reaction
DNA samples were extracted using the i-genomic Plant DNA Extraction Kit (iNtRON Biotechnology, Seongnam, Korea) following the manufacturer’s instructions. Polymerase chain reaction (PCR) was performed using specific primers for each gene with Takara Ex Taq (Takara Bio Inc., Otsu, Japan) and Quick Taq HS DyeMix (Toyobo, Osaka, Japan). All the reactions were performed in 50 μL volumes containing 5 μL 10× Ex Taq buffer, 4 μL 2.5 mM each dNTP, 4 μL each of the appropriate primers at 10 pM, and 0.3–0.5 μL Taq DNA polymerase (Takara Ex Taq). PCR cycle and amplification were performed with SSU-rDNA primer pairs (G01/G14, G04/G07) (Saunders and Kraft 1994) and the rbcL primer pairs (FrbcL/R1150, F753 or F993+/RrbcSp) (Freshwater and Rueness 1994, Bárbara et al. 2013). PCR products were purified with the QIAquick PCR Purification Kit (Qiagen Inc., Valencia, CA, USA) or the MG PCR Product Purification Kit (Microgen, Seoul, Korea) following the manufacturer’s recommendations for direct sequencing. Sequencing reactions were performed with the BigDye Terminator Cycle Sequencing Kit (Macrogen, Seoul, Korea), and samples were loaded in an ABI 3730 DNA Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). Sequence reads were manually assembled and edited in BioEdit ver. 7.0.5 (Hall 1999), and chromatograms were checked to confirm the validity of ambiguous nucleotides.
Phylogenetic analysis
A dataset consisting of 104 rbcL sequences (including our six newly determined sequences [MG926655-MG926676] and three outgroups were trimmed to the same length of 1,313 bp) and were used to evaluate the phylogenetic position of our newly acquired Pyropia species. We selected one sequence from each currently listed species of Pyropia and putative cryptic taxa from GenBank (Supplementary Table S1) (Guiry and Guiry 2017) to provide broader a distributional pattern for the Pyropia genetic group as possible. However, short rbcL sequences (ca. 870 bp) of putative cryptic species of Pyropia from Chile (López-Vivas et al. 2015, Guillemin et al. 2016) were discarded prior to the final analysis. In addition, five species and one variety, which are currently listed taxa in AlgaeBase (Guiry and Guiry 2017) were excluded because of the lack of available sequence data (e.g., P. drachii) or short rbcL sequences (P. orbicularis, P. raulaguilarii). The registered rbcL sequences of P. pulchella (Ackland, J. A. West, J. L. Scott & Zuccarello) T. J. Farr & J. E. Sutherland, P. stamfordensis C. Neefus, T. Bray & A. C. Mathieson (as named Porphyra sp. stamfordensis), and P. collinsii C. Neefus, T. Bray et A. C. Mathieson (as named Porphyra sp. collinsii) in GenBank were also excluded because the sequence of P. pulchella the same of P. kuniedae, and P. stamfordensis and P. collinsii the same of P. parva (see Supplementary Table S1).
Appropriate models of sequence evolution were assessed using jModelTest 2 (Darriba et al. 2012) under both AIG criteria. Model selection occurred with the following parameters: GTR + I + G with gamma distribution = 1.4330; proportion of invariable sites = 0.6340; base frequencies A = 0.3222, C = 0.1460, G = 0.1698, T = 0.3620; and rate among sites [A–C] = 0.7479, [A–G] = 5.7755, [A–T] = 0.5708, [C–G] = 0.4281, [C–T] = 8.8906, and [G–T] = 1.0000. Maximum likelihood analyses were conducted using PhyML ver. 3.0 (Guindon et al. 2010), under the appropriate model of sequence evolution, and with support both calculated by an approximate likelihood ratio test (aLRT) (Anisimova and Gascuel 2006) and by a bootstrap analysis using 1,000 pseudoreplicates. Bayesian inference was implemented in MrBayes ver. 3.1.2 (Huelsenbeck and Ronquist 2001). Two independent analyses were run with four chains of the Markov chain Monte Carlo (one hot and three cold), sampling one tree every 100 generations for 1,000,000 generations, starting with a random tree. A total of 30,000 generations were discarded as “burn in.” p-Distance analyses were calculated for the phylogenetic matrix in MEGA ver. 5.05 (Tamura et al. 2011).
RESULTS
Molecular analysis
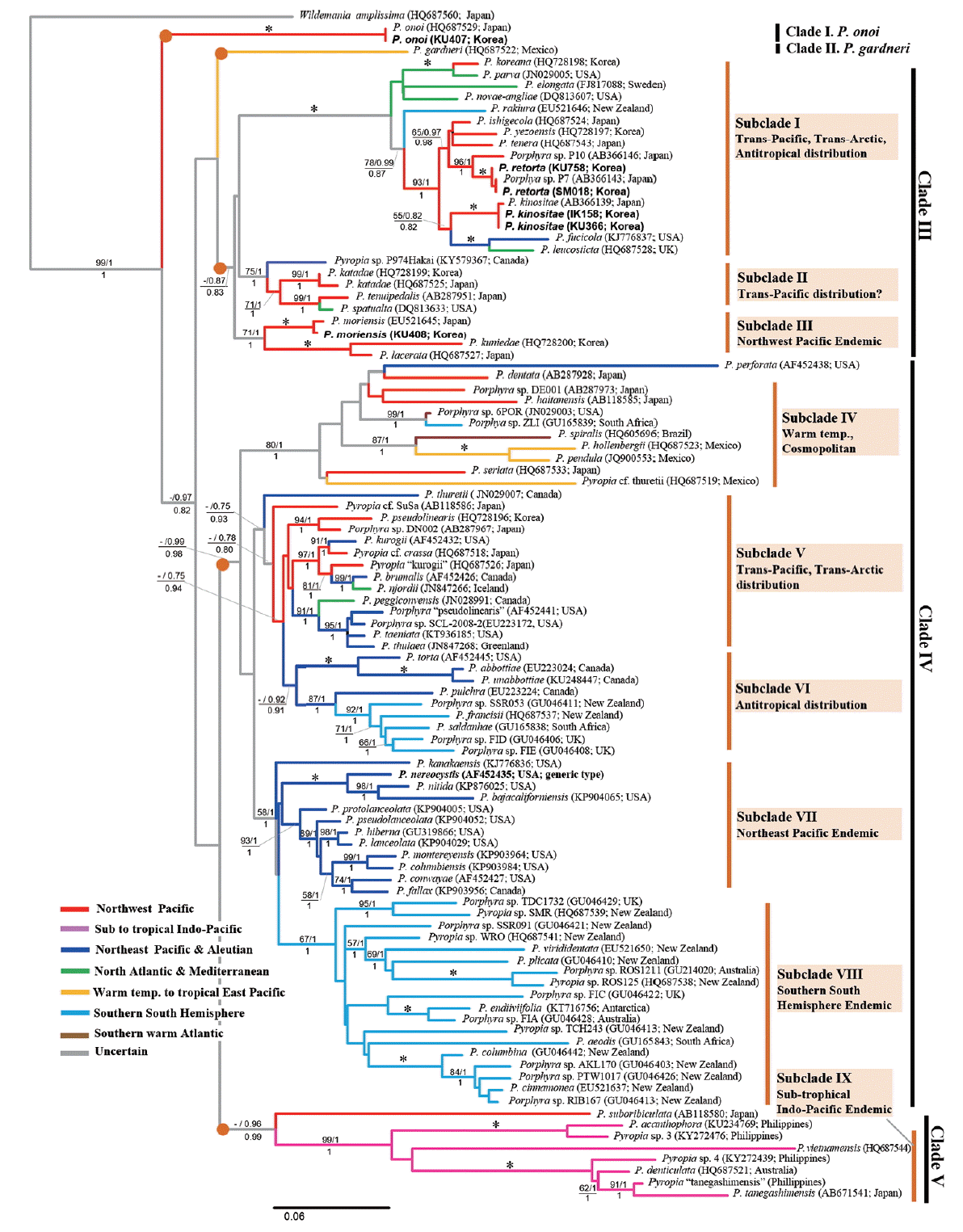
In our reconstructed maximum likelihood (ML) phylogram (−lnL = 12,761.2691) (Fig. 1), the species of the genus Pyropia formed a well-supported monophyletic taxon, as in previously published trees of rbcL (Kucera and Saunders 2012) or concatenated rbcL-rSSU trees (Sutherland et al. 2011, Meteo-Cid et al. 2012, Sánchez et al. 2014). Topologies of our data were similar with previously reconstructed trees (Fig. 1), but slightly differed from those of ML and Bayesian trees by the position of P. garderni, which was nested as sister of Clade V in Bayesian analysis (data not shown). Our rbcL ML tree was resolved with five moderately supported clades: 1) P. onoi clade; 2) P. gardneri clade; 3) Clade III [supporting values (sv): ML bootstrap / Bayesian posterior probability / aLRT value (sv: −/0.87/0.83)]; 4) Clade IV (sv: −/0.78/0.80); and 5) Clade V (sv: −/0.96/0.99). The early separated clade corresponding to P. onoi was similar to that of previous reports (Sutherland et al. 2011, Sánchez et al. 2014). Each clade had a tendency for a biogeographic pattern; these five clades are divided into approximatively nine subclades based on clustering and distribution (Fig. 1).
Interestingly, the putative endemic species of the Northwest Pacific were scattered subclades in I, VI, and V, and mixed with the Northeast Pacific or Arctic to North Atlantic species. Moreover, strong sibling relationships between the Northwest Pacific and the Northeast Pacific species or between the Northeast Pacific and Arctic or North Atlantic species (e.g., P. kinositae-P. fucicola-P. leucosticta and P. kurogii-P. brumalis-P. njordii) were convincing evidence of a link between species diversification of Pyropia and the Trans-Pacific to Trans-Arctic expansion histories from the Northwest Pacific members. Our phylogenetic tree also showed evidence of anti-tropical distribution relationships between the Northwest Pacific and the southern hemisphere species in subclades I and VI and between subclades VII and VIII. On the other hand, four strict biogeographic endemic clades were resolved: in the Northwest Pacific (subclade III), in the Northeast Pacific (subclade VII), in the southern South hemisphere (subclade VIII), and in the Indo-Pacific waters (subclade IX).
Delineation of species based on rbcL sequence data
Our 24 newly determined complete rbcL sequences from Korean specimens (Table 1) identified four species, P. kinositae, P. moriensis, P. onoi, and a cryptic species (KU758 and SM018) (Fig. 1). Twenty-four sets of sequence data among them were nested in the subclade I at two terminal branches (Fig. 1). The subclade was well resolved as a strongly supported monophyletic clade, which was composed of 13 taxa at the species level, including two of the most economically important species for aquaculture, P. yezoensis (Ueda) M. S. Hwang & H. G. Choi and P. tenera (Kjellman) N. Kikuchi, M. Miyata, M. S. Hwang & H. G. Choi (Fig. 1). One type of rbcL sequence (8 out of 24) in our data was identified as P. kinositae (Yamada & Tak. Tanaka) N. Kikuchi, M. Miyata, M. S. Hwang & H. G. Choi, which had previously only been reported in Northern Japan (Hokkaido and the northeast coast of Honshu) based on the literature and GenBank data, and another type of rbcL sequence (14 out of 24) was identical with those of Porphyra “P7” (GenBank accession No. AB366143). The other two rbcL sequences (KU407 and KU408) were nested in Japanese P. onoi (Ueda) N. Kikuchi & M. Miyata and P. moriensis (Ohmi) N. Kikuchi & M. Miyata, respectively.
Taxonomic treatment
Pyropia retorta sp. nov. S. -M. Kim, H. -G. Choi & H. -S. Kim
Description
Thalli membranous, monostromatic, color darkish red in the upper portion and greenish red in lower the portion, axially curled or twisted, shape lanceolate to oblanceolate with obtuse to rotund base without conspicuous stipe (Fig. 2A–C), 6–15 (−35) cm long, 0.8–3 (−8) cm broad, and 35–42 μm thick in the central vegetative portion. Margins entire, slightly undulated (Fig. 2D). Each cell contains a single stellate plastid with central pyrenoid. Vegetative cells oblong or irregular tetragonal to polygonal with rounded angles, and 15–25 μm long × 8–15 μm broad in surface view, and oblong and quadrate with round angles, 20–23 μm high × 8–15 μm broad in sectional view (Fig. 2F & G). Basal cells capitate with projected rhizoidal filaments, 15–40 μm long × 10–15 μm broad in surface view. Rhizoidal filaments arranged in both directions in sectional view. Thalli mixed monoecious. Spermatangia in longitudinal linear patches with a sharp boundary, formed and arranged in parallel with zygotosporangial patches distally and along margins, each containing 128 (a/4, b/4, c/8) at maximum (Fig. 2C, J & K). Prototrichogyne conspicuous, acute to obtuse (Fig. 2H). Zygotosporangia containing 16 (a/2, b/2, c/4) at maximum (Fig. 2I & J). Thalli 47–50 μm thick in spermatangial portion, 58–62 μm in zygotosporangial portion.
Holotype
Gametophyte NIBRRD0000003217 (SM018) collected on Mar 20, 2004 in Jumunjin, Gangneung (Gangwon-do, Korea; 37°54′21.80″ N, 128°49′49.77″ E) by S. M. Kim and deposited in the National Institute of Biological Resources (NIBR), Incheon, Korea.
Representative specimens examined
IK111, KN033, KU652, KU654, KU655, KU656, KU668, KU757, KU758, KW020, and SM018 deposited in the Herbarium of Gangneung National University (Table 1).
Habitat and geographical distribution
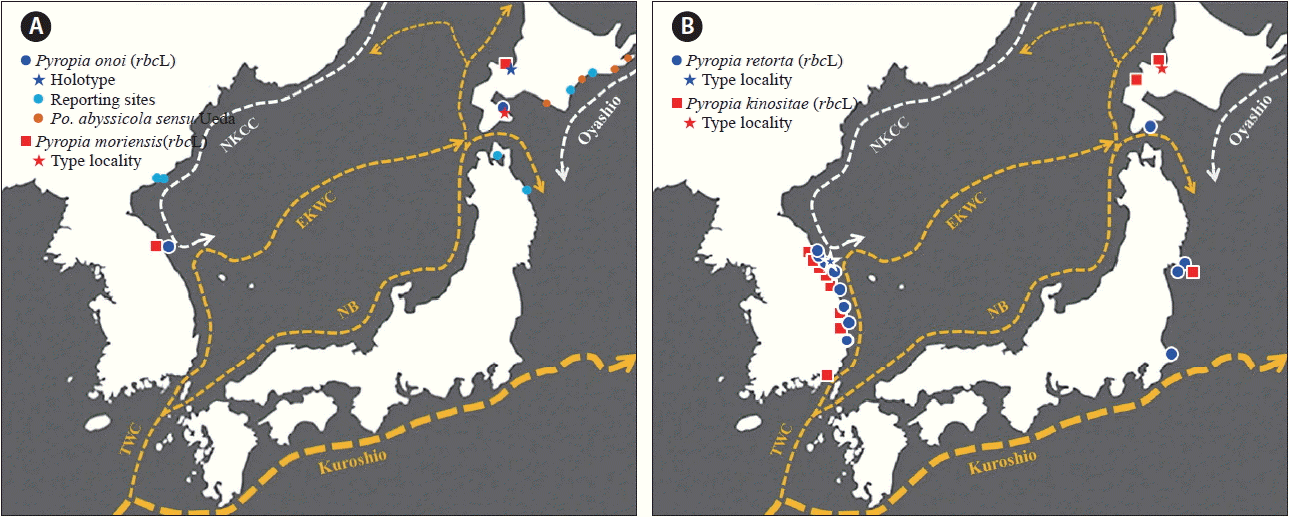
Blade phase is epilithic, inhabiting exposed intertidal areas, where splashed intermittently by waves. This phase is a seasonal annual, occurring from January to April, and it can form dense populations solely or intermixed with P. pseudolinearis (Ueda) N. Kikuchi, M. Miyata, M. S. Hwang & H. G. Choi. To estimate the distribution range based on GenBank data and our molecular work, this species is only known in the Northwest Pacific, as an endemic, from the east coast of Korea to Northern Japan (Hokkaido, northern east coast of Honshu) (see Fig. 3).
Morphology
The thalli are heavily twisted or axially curled in nature, and thus very difficult to spread on herbarium sheets to make dry specimens (Fig. 2B). Thalli are darkish red in the upper part and greenish red in the lower part when fresh (Fig. 2B & C), and darkish brown with a yellowish green basal portion when pressed and dried at first (Fig. 2A & B), but the greenish color gradually fades as time goes in herbarium specimens (Fig. 2A). Specimens adhere moderately well to paper. This species is always monoecious with conspicuous linear and narrow (400–550 μm broad) spermatangial patches, which are almost parallel to the axis and penetrated by broaden darkish red zygotosporangial areas of the upper blade margins (Fig. 2C). Archeospores (monospores) were not observed, at least in wild plants.
DISCUSSION
Taxonomic perspective
Pyropia is the most speciose genus among bladed Bangiaceae, and has a wide geographic distribution around the world, from tropical to subpolar waters (Sutherland et al. 2011). Although 70 species names are currently accepted taxonomically in the genus Pyropia (Guiry and Guiry 2017), three species among them were not resolved as distinct species in our rbcL phylogenetic analysis. Pyropia collinsii Neefus, T. Bray & A. C. Mathieson should be treated as a heterotypic synonym of Pyropia parva A. Vergés & N. Sánchez based on identical rbcL sequence data between both species (see Sánchez et al. 2014). On the other hand, P. pulchella (Ackland, J. W. West, J. L. Scott & Zuccarello) T. J. Farr & J. E. Sutherland and P. stamfordensis Neefus, T. Bray & A. C. Mathieson should be treated as heterotypic synonyms of P. kuniedae (Kurogi) M. S. Hwang & H. G. Choi based on identical rbcL sequences among them.
The occurrence of P. onoi is confirmed based on molecular data in Korea after being reported from the northern part of the east coast of North Korea (Ueda 1932). The specimens were collected only once during this study as epiphytes on Grateloupia asiatica S. Kawaguchi & H. W. Wang. In the field, the epiphytic character of this species attracted our attention even though its gross morphology is similar to epilithic P. suborbiculata (Porphyra okamurae sensu Ueda), which is abundant at the collection site. Under microscopic observations, the specimens of the former are clearly distinguished from the latter by the entire margin of thallus, monoecious thalli with narrow spermatangial streaks around the margin, and absence of monospore production (Supplementary Fig. S1). P. onoi has been known as an epiphyte on Mazzaella japonica (Mikami) Hommersand and Chondrus yendoi Yamada & Mikami (Ueda 1932). Based on the rbcL sequence data, the Korean specimen has the same haplotype as that reported from Hokkaido, Japan (HQ687529).
Also, the distribution of P. moriensis on the east coast of Korea was confirmed for the first time in this study. However, we could collect only one specimen, which was cast ashore, in spite of several attempts and much attention to obtain more material. The specimen was mixed with numerous cast ashore thalli of P. kinositae, but it was distinguished from the latter by its pale reddish color and absence of longitudinal linear spermatangial patches (Supplementary Fig. S2). This species has been known as an obligate epiphyte on Chorda filum (Linnaeus) Stackhouse (Ohmi 1954, Notoya and Miyashita 1999); however, the host of the Korean specimen was indeterminable because the cast ashore thallus was detached. C. filum is relatively common on the south and southwest coast, but rare on the east coast of Korea, only collected once when it was cast ashore on the northern east coast (personal observation). The rbcL sequences of the Korean and Japanese specimens differed by 2 bp substitutions.
Our rbcL data also confirmed the widespread distribution of P. kinositae on the east coast of Korea (Fig. 3B). Most specimens were collected when they were cast ashore from late January to the middle of May on the east coast of Korea, especially in Gangwon-do Province. We could verify the habitat of this species by SCUBA diving collection in shallow to moderately deep subtidal zone (ca. to 20 m depth) on bare rocks or on coralline algae. The species was described as a forma as Porphyra yezoensis f. kinositai (Tanaka 1952), largely based on its subtidal habitat (3–7 fathoms = ca. 5–15 m in depth). The darkish purplish red thalli are one of the diagnostic characters distinguishing this species from P. yezoensis, which is greenish in color at least in the lower part of the thallus (Kim and Kim 2011). The other morphological characteristics of the Korean samples agreed well with the descriptions of Tanaka (1952) and Fukuhara (1968) (Supplementary Fig. S3). All rbcL sequences of Korean specimens were identical with those of Japan specimens (AB366145, EU521641), except for 1 bp substitution in the Japanese Otaru specimen (AB366139) among the 1,467 bp of the complete rbcL sequence. The species has not been recorded on the west coast of Japan to date and is distributed only on the west and southern coasts of Hokkaido and the northern part of Honshu (Fukuhara 1968) (Fig. 3B).
Another epilithic Pyropia sp. was shown to be having a similar biogeographic pattern to that of P. kinositae (Fig. 3B). The rbcL sequences of 14 Korean specimens were identical or differed by only 1 bp substitution with those of Japanese Porphyra sp. P7 (AB366143). This species is common and usually collected from the lower tide level to the supralittoral zone (ca. 1 m level up from the high-tide level) in the east coast of Korea. The occurrence of this cryptic species was previously recognized in Korea based on molecular data by Kim (2005) and Niwa et al. (2014). Furthermore, Niwa and Kobiyama (2014) verified the interspecific hybridization between Pyropia sp. 2 (= P. retorta) and Pyropia sp. 3 (Pyropia sp. P10 in Fig. 1) as in the case of P. yezoensis and P. tenera (Niwa et al. 2009, Niwa and Sakamoto 2010). This species is clearly distinguished by its unique morphological characteristics and rbcL sequences from the closely related species P. yezoensis, and thus we proposed as a new species P. retorta in this study.
Biogeographic pattern of the four native Pyropia
Our newly determined rbcL sequence data can be used to describe the biogeographic patterns of the four native species. The four species are basically restricted to the east coast of Korea and the western to southern part of Hokkaido (and northeastern Honshu) in Japan. The biogeographic pattern of the two species, P. kinositae and P. retorta, suggests that they are native to the east coast of Korea, and it seems highly probable that they were isolated and remained in the southern part of East Sea (Sea of Japan) during the Last Glacial Maximum (LGM): the pattern coincides with the East Korean Warm Current (Fig. 3B). Although the distributional data is limited to date, P. onoi and P. moriensis show different pattern that relates with North Korean Cold Current because they are not observed in the southern east coast of Korea (Fig. 3A). However, the distribution range of P. onoi should be revised based on molecular data including the representatives of Porphyra abyssicola sensu Ueda (1932) in Japan. The distribution of the former two species, P. kinositae and P. retorta, may have expanded to northern Japan through the restored East Korean Warm Current after the LGM (Fig. 3B). Interestingly, Pyropia sp. P10 (AB366146), considered a sibling species of P. retorta currently, has not been found on the east coast of Korea to date in spite of our close attention in collection. The distributional differences between P. retorta and Pyropia sp. P10, although currently sympatric in the northeast coast of Japan (Niwa et al. 2014), suggests that they may be a vicariant pair, possibly separated relicts from different refuges; one located on the east coast of Korea (P. retorta) and the other perhaps placed on the east coast of central Japan (Pyropia sp. P10) during the LGM. The scenario of postglacial dispersal from different refuges, however, should be reconfirmed with further phylogeographic research based on more rapid evolutionary genetic markers than rbcL.
Eight native species of Pyropia occur on the east coast of Korea: 1) P. kinositae; 2) P. koreana (M. S. Hwang & I. K. Lee) M. S. Hwang, H. G. Choi, Y. S. Oh & I. K. Lee; 3) P. moriensis (Ohmi) N. Kikuchi & M. Miyata; 4) P. retorta; 5) P. onoi (Ueda) N. Kikuchi & M. Miyata; 6) P. pseudolinearis (Ueda) N. Kikuchi, M. Miyata, M. S. Hwang & H. G. Choi; 7) Porphyra akasakae Miura; and 8) Po. irregularis Fukuhara (Kim and Kim 2011, This study). These species have only been reported in northern Japanese waters (Hokkaido, northeastern Honshu) or the east coast of Korea and the west coast of Japan, but not along the south coast of Korea (except P. kinositae in this study). The distribution pattern of these eight species suggests that the East Sea (= Japan Sea) seems to have played a major role as refugia during the LGM.
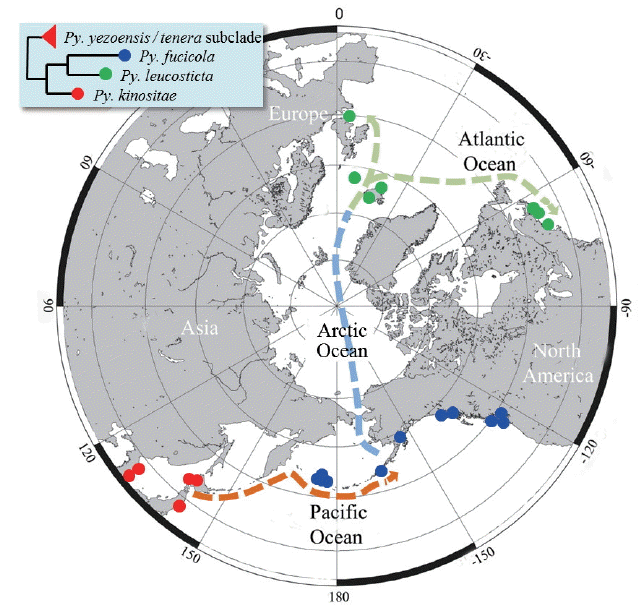
It has long been recognized that there are two kinds of endemic species: neo-endemics–recently diverged species that are endemic because of lack of dispersal / migration out of their ancestral area; and paleo-endemics–old species that were perhaps more widespread in the past and are now restricted to a local region (Nekola 1999, Mishler et al. 2014). It could be a reliable method to peer into the hypothesis of biogeography through the species phylogenetic tree combined with distributional information, and also to detect the differentiation between areas with clusters of new emerging species (neo-endemics) and areas with clusters of unique, but disappearing, species (paleo-endemics) that often occur in restricted areas of their refuges or origin (Mishler et al. 2014). Following the concept of endemics, P. retorta is a neo-endemic, whereas P. kinositae is a paleo-endemic species in our phylogenetic tree (Fig. 1) and as shown in the scenario of the historical biogeography of the P. kinositae lineage (Fig. 4).
Interestingly, an accelerated rate of evolution was found in subclades IV, VIII, and IX, which have remarkably long branches leading to terminal species. Generally, an accelerated rate of evolution may be caused by an increased fixation rate of new mutations, which is affected by natural selection (purifying), population size, and rate of cladogenesis (Mindell and Thacker 1996). The relatively milder environments in the Southern hemisphere and warm temperate to tropical Oceans compared to those of the northern Pacific during the glacial epoch probably led to the acceleration, which may have been caused by weak selection pressures and few cladogenesis or fragmentation of populations. On the other hand, the rapid cladogenesis of Pyropia that was recognized on both sides of the North Pacific may occur because of Trans-Pacific dispersal and repeated fragmentation of populations in response to climatic cooling during the Cenozoic (Briggs 2003). The Trans-Pacific and / or Trans-Arctic dispersal followed by long-term isolation (and bottlenecks?) was observed in the P. fucicola / P. leucosticta / P. kinositae lineage as shown in the scenario of the historical biogeography of the Pyropia kinositae lineage (Fig. 4), and in the P. kurogii / P. njordii and P. peggiconvensis / Porphyra sp. SCL-2008-2 lineage (Fig. 1). Generally, the subtropical to warm temperate species of Pyropia, e.g., P. acanthophora show much more genetic diversity including 20 rbcL haplotypes (Dumilag and Aguinaldo 2017). However, P. kinositae and P. retorta have low genetic diversity, with only two rbcL haplotypes detected. The low genetic diversity of these two species may be the result of a severe bottleneck effect (purifying) during the glacial epoch in the Cenozoic or recent population expansion.
Much attention is needed to explain the hypothesis of biogeography of the well resolved subclade I, which is composed of 13 species including P. yezoensis, an economically important species for nori cultivation in Korea and Japan (Fig. 1). It is thought that members of this subclade have complex dispersal histories, including Trans-Pacific, Trans-Artic, and Trans-equatorial (anti-tropical) distributions. This subclade is a monophyletic lineage with a very strongly supported branching pattern in the rbcL phylogeny (Kucera and Saunders 2012, This study) and SSU-rDNA + rbcL concatenated data (Sutherland et al. 2011, Sánchez et al. 2014). The long ancestral branch of this genetic lineage likely reflects an earlier separation with long independent history of cladogenesis from the sister subclades II and III. Overall, a complex dispersal hypothesis, including Trans-Pacific, Trans-Arctic, and Trans-equatorial can be safely stated to explain the cladogenesis of this genetic lineage. The hypothesis of Trans-Pacific and / or Trans-Arctic migration has been well-documented in many marine biota, especially focusing on the North Pacific–North Atlantic connections during the Pleistocene based on molecular data (Lindstrom 2001, Nikula et al. 2007, Coyer et al. 2011). Trans-equatorial dispersal based on molecular data has been discussed in the case of marine algae (Hommersand and Fredericq 2003), for some marine animals (Williams et al. 2003, Mabuchi et al. 2004, Gérard et al. 2008, Naughton et al. 2014) and terrestrial plants (Nakamura et al. 2012).