ABSTRACTCyanobacteria have been widely reported to produce a variety of UV-absorbing mycosporine-like amino acids (MAAs). Herein, we reported production of the unusual MAA, mycosporine-glycine-alanine (MGA) in the cyanobacterium Sphaerospermopsis torques-reginae ITEP-024 using a newly developed UHPLC‐DAD‐MS/HRMS (ultra-high performance liquid chromatography–diode array detection–high resolution tandem mass spectrometry) method. MGA had previously been first identified in a red-algae, but S. torques-reginae strain ITEP-024 is the first cyanobacteria to be reported as an MGA producer. Herein, the chemical structure of MGA is fully elucidated from one-dimensional / two-dimensional nuclear magnetic resonance and HRMS data analyses. MAAs are unusually produced constitutively in S. torques-reginae ITEP-024, and this production was further enhanced following UV-irradiance. It has been proposed that MAA biosynthesis proceeds in cyanobacteria from the pentose phosphate pathway intermediate sedoheptulose 7-phosphate. Annotation of a gene cluster encoded in the genome sequence of S. torques-reginae ITEP-024 supports these gene products could catalyse the biosynthesis of MAAs. However, addition of glyphosate to cultures of S. torques-reginae ITEP-024 abolished constitutive and ultra-violet radiation induced production of MGA, shinorine and porphyra-334. This finding supports involvement of the shikimic acid pathway in the biosynthesis of MAAs by this species.
INTRODUCTIONCyanobacteria are photosynthetic prokaryotes that probably evolved during the Precambrian era (around 2.9 Ga-) to create the oxygenic environment that supports current life of Earth (Fischer 2008, Rastogi et al. 2014). Cyanobacteria harness the sun’s photosynthetically active radiation (PAR) to convert light into chemical energy. An obligate requirement for PAR results in prolonged exposure to ultra-violet radiation (UVR) which can directly cause cellular damage due to photochemical reactions such as dipyrimidine photodimers in DNA, or indirectly via the accumulation of reactive oxygen species (Rastogi et al. 2010). In order to circumvent the harmful effects associated with exposure to UVR, cyanobacteria and other aquatic organisms have evolved biochemical and mechanical defences (Shick and Dunlap 2002, Singh et al. 2008b, Rastogi et al. 2019). Among these is the ability to synthesize, or otherwise acquire, mycosporine-like amino acids (MAAs). MAAs are small (<400 Da), colourless, water soluble metabolites, comprised of either an aminocyclohexenimine or aminocyclohexenone UV-absorbing chromophore, conjugated to a nitrogen or amino-alcohol component (Shick and Dunlap 2002). The absorption maxima of MAAs (λmax) are within either ultraviolet B (UVB) and ultraviolet A (UVA) spectra of solar radiation (Cockell and Knowland 1999), and MAAs have been convincingly demonstrated to possess physiologically relevant photo-protective and antioxidant properties (Lawrence et al. 2018). Thus, MAAs have been widely studied as natural alternatives to potentially toxic synthetic sunscreens and for other biotechnological applications (Lawrence et al. 2017). We undertook a screening program to identify novel MAAs from cyanobacteria deposited in the culture collection of the Laboratory of Toxins and Natural Products of Algae and Cyanobacteria (LTPNA) at the University of São Paulo (Geraldes et al. 2019). S. torques-reginae strain ITEP-024 is a filamentous and heterocystous cyanobacterium that normally occurs as dense freshwater blooms and in waters with low salinity throughout South America (Molica et al. 2005, Werner et al. 2012). Few secondary metabolites have been isolated from this strain, such as anatoxin-a(s), namalides, spumigins (Dörr et al. 2010, Sanz et al. 2015, 2017) while the biosynthesis of MAAs has never been described. Herein, the production of three MAAs by S. torques-reginae ITEP-024 is reported, including mycosporine-glycine-alanine (MGA), which was isolated for the first time from a cyanobacterium (Supplementary Fig. S1). Molecular biology studies, UVR induction experiments and the use of a shikimate pathway inhibitor (glyphosate) afforded biosynthetic insights about MAAs production by the strain.
MATERIALS AND METHODSReagents and matrixAcetonitrile (J. T. Baker), formic acid, and ammonium formate were liquid chromatography–mass spectrometry grade (Fluka, Seelze, Germany). Water was purified by a Milli-Q system (Millipore, Milford, MA, USA). Porphyra-334 and shinorine standards were kindly provided by Prof. Kazuo Yabe. Glyphosate (96%) was purchased from Sigma-Aldrich (St. Louis, MO, USA).
Cyanobacterial cultures
Sphaerospermopsis torques-reginae strain ITEP-024 was isolated from a toxic bloom sample collected in the Tapacura Reservoir, Recife, Brazil (Molica et al. 2005). S. torques-reginae ITEP-024 was cultivated in ASM-1 medium (Gorham et al. 1964) at 24 ± 2°C under a light : dark cycle (12 h : 12 h) with PAR supplied by OSRAM L 36 W : 32 Lumilux de luxe warm white lamps (80 μmol m−2 s−1).
MAAs extraction and partial purificationCyanobacterial cells were harvest after 15 days growth by centrifugation at 10,000 ×g for 15 min at 5°C and lyophilized. Then, cells were extracted with 0.2% (v/v) formic acid solution + 2 mM ammonium formate. Sample were mixed by vortex and stored for 1 h at room temperature. No ultrasonic treatment of the lyophilized cyanobacteria or any other approach to lyse or homogenize the cells were used. Cell debris was removed by centrifugation at 10,000 ×g for 15 min and the supernatant was retained as the partially purified MAA fraction.
MAAs isolation via high performance liquid chromatography–diode array detection (HPLC-DAD)MAAs were purified by semi-preparative Shimadzu Prominence liquid chromatograph-LC 20AT equipped with a photodiode array detector DAD-SPD M20A (Shimadzu, Kyoto, Japan) and a fraction collector fitted to a Luna C18 (2) column (250 × 10 mm, 5 μm; Phenomenex, Torrance, CA, USA). Buffer A was 0.2% (v/v) formic acid solution + ammonium hydroxide (pH 3.15), buffer B was Milli-Q water/acetonitrile/methanol (8 : 1 : 1) + formic acid (pH 2.20), buffer C was acetonitrile/methanol (1 : 1) + 0.1 % (v/v) formic acid and the flow rate was 0.5 mL min−1. The gradient used for separation was 100% A at the start of the run, 22% B at 6 min, 95% C at 13 min, where the gradient was held for 10 min, followed by a 10 min equilibration at 100% A prior to the next injection. Compound (3) was collected at 13.0 min in the run time (Supplementary Fig. S2).
UHPLC-DAD-MS/HRMS analysisThe UHPLC-DAD-MS/HRMS (ultra-high performance liquid chromatography-diode array detection-high resolution tandem mass spectrometry) analyses were carried out on a Shimadzu Prominence system (Shimadzu) coupled to a quadrupole time-of-flight mass spectrometer (MicroTOF-QII; Bruker Daltonics, Billerica, MA, USA) with an electrospray source. Sample extracts were injected (10 μL) onto a Synergi 4 μ Hydro-RP 80 A column (150 × 2.0 mm) (Phenomenex, Torrance, CA, USA) and eluted at a flow rate of 0.4 mL min−1 with 0.1% (v/v) formic acid solution + 2 mM ammonium formate (buffer A). Eluent B was acetonitrile/Milli-Q water (8 : 2) + 0.1% (v/v) formic acid. The gradient used for the separation was 100% A at the start of the run up 2 min, 95% B at 6 min, where the gradient was held for 1.5 min, followed by a 4 min equilibration at 0% B prior to the next injection. The ionization source conditions were as it follows: positive ionization, capillary potential of 4,500 V, temperature of drying nitrogen gas 200°C at a flow rate of 9 mL min, nebulizer pressure of 60 psi. Mass spectra were acquired using electrospray ionization in the positive mode over a range of m/z from 50 to 500. The QTOF instrument was operated in scan and Auto tandem mass spectrometry (MS/MS) mode, performing MS/MS experiments on the three most intense ions from each MS survey scan. The chromatogram was monitored at 330 nm (indicative of the MAAs shinorine and porphyra-334) and retention times of eluted peaks were compared shinorine and porphyra-334 standards (kindly provided by Prof. Kazuo Yabe). Accurate mass data were processed using Data Analysis 4.0 software (Bruker Daltonics).
NMR analysisOne-dimensional (1D) and two-dimensional (2D) nuclear magnetic resonance (NMR) spectra were obtained using a 500 MHz (125 MHz for 13C) Bruker DPX spectrometer. NMR experiments were run using standard pulse sequences. Chemical shifts are reported on the δ scale relative to the residual solvent signal (CD3OD).
Gene cluster identification and annotationPrediction of genes encoding putative MAA biosynthetic enzymes was performed by mining the draft S. torques-reginae ITEP-024 genome sequence (Lima et al. 2017) using TBLASTN (Boratyn et al. 2013). Artemis 18.0.2 (Carver et al. 2011), BLASTP and HHpred (Zimmermann et al. 2018) searches against the GenBank database (Sayers et al. 2019) were used for manual annotation of putative gene clusters.
MAAs induction using UVRTo induce MAA biosynthesis, cells were irradiated using a UVA source (maximum illumination at 370 nm) provided by twin Philips TL-K 40W/10-R UV-A lamps, and a UVB source (maximum illumination at 290 nm) provided by twin Philips TL 40W/12 RS SLV/25 lamps in addition to the PAR lamps (maximum illumination at 550 nm). Cell density were normalized to 0.4 mg dry weight mL culture−1, and 30 mL of suspended cells were irradiated in 25 cm2 tissue culture flasks with a hydrophobic filter cap (TP90026, TPP) at a distance of 40 cm with lamps at 24 ± 2°C under light / dark cycle (12 h : 12 h) for 3 days (Singh et al. 2008a). Cultures were harvest at the beginning and at the end of the experiment, and MAAs were extracted for subsequent analysis of MAAs using the liquid chromatography-tandem mass spectrometry (LC-MS/MS) method described by Geraldes et al. (2020). All the experiment was performed in triplicate.
MAAs induction in the presence of glyphosateCells were grown with illumination for 15 days at 24 ± 2°C under a light : dark cycle (12 h : 12 h) with PAR lamps (80 μmol m−2 s−1), before the culture medium was supplemented with 2 mM glyphosate. Growth of the culture was allowed to proceed for a further 24 h to allow cells to absorb glyphosate before exposure to UVR (Shick et al. 1999). Then, MAAs were induced for 3 days as described above (n = 3).
RESULTS AND DISCUSSIONDistribution of MAAs in Sphaerospermopsis torques-reginae strain ITEP-024A UHPLC-DAD-MS/HRMS based protocol was implemented to screen for photoprotective compounds in cyanobacteria from the LTPNA culture collection. Interestingly, analysis of biomass extracts from S. torques-reginae ITEP-024 indicated the detection of three target compounds, that eluted at 1.3, 2.2, and 3.1 min (Supplementary Fig. S3), which showed diagnostic chemical features for MAAs such as low retention time in reversed phase, and characteristic UV and MS/MS spectra profiles. The corresponding molecular formulae C13H20N2O8 (1), C14H22N2O8 (2), and C13H20N2O7 (3) were calculated from the protonated ions [M + H]+ to m/z 333.1298 (0.0 ppm), m/z 347.1451 (−0.9 ppm), and m/z 317.1350 (0.3 ppm) (Supplementary Fig. S4). Noteworthy, all compounds showed loss of a methyl radical, followed by sequential CO2 eliminations (Cardozo et al. 2006) according to the detected common losses of 15 Da and 44 Da (Supplementary Fig. S5). All UV spectra were very similar, suggesting the same conjugated chromophore with a maximum absorption at 332 nm. Accurate masses were then used as databases queries to search the Dictionary of Natural Products (Taylor and Francis Group 2017), Antibase (Laatsch 2014), and MAA literature. Shinorine and porphyra-334 were suggested as possible MAAs for compounds (1) and (2). The chemical identity of these compounds was then confirmed using UV-HRMS data analyses in comparison to authentic reference standards. MGA identity was suggested for compound (3). This MAA is a rare mycosporine previously reported by Orfanoudaki et al. (2019) as a constituent of red algae and as a shunt product during MAA biosynthesis in a heterologous host (Miyamoto et al. 2014). However, MGA had never been reported in a cyanobacterial strain until recently by our group (Geraldes et al. 2020).
Isolation and characterization of compound (3)Biomass extract of S. torques-reginae ITEP-024 was separated by reversed phase chromatography, allowing the isolation of MGA (3). The chemical structure of (3) was established by 1D and 2D NMR, and high resolution tandem mass spectrometry (MS/HRMS) data analysis. The 1H-NMR spectrum of (3) revealed the presence of one methyl group, four methylenes, one methyl and one methoxy group. The COSY spectrum defined three spin systems including the coupling of the pairs of duplets from the diasterotopic methylenes at δH 2.65/2.88 (CH2-4) and δH 2.73/2.95 (CH2-6) (Fig. 1A) showing a geminal constant coupling (J = 17 Hz for both), as well as the nuclei coupling from methyl group at δH 1.48 (CH3-13) with methine protons at δH 4.16 (CH-11), indicating the evidence of an alanine moiety for (3). The methyl protons at δH 1.48 (CH3-13) were also confirmed through the long-range correlation detected in the heteronuclear multiple-bond correlation spectrum (Fig. 1B) with the carboxylic carbon (COOH-12) at δC 173 and the carbon nucleus attached to the imino moiety (C-11) at δC 55. Moreover, 2J and 3J-bond correlations from δH (H-7) to δC (C-5) and δC (C-6) were detected, supporting the oxymethylene protons; while the 3J-bond correlation from δH 3.66 (H-8) to the sp2 carbon (C-2) at δC 122 allowed the assignment of the methoxy group attached to the aminocyclohexenimine ring.
Full NMR assignments are described in Table 1 and are in agreement with data reported for MGA produced by heterologous expression (Miyamoto et al. 2014). The 1D and 2D spectra with main nuclei correlations are depicted in Supplementary Figs S6–S11.
MS/HRMS data analysis also confirmed the structure of (3). The spectrum of (3) (Supplementary Fig. S5) displayed the characteristic fragmentation pattern of a di-acidic MAA in positive ionization mode (Cardozo et al. 2009). In particular, as a first fragmentation step the loss of a methyl radical from a protonated molecular ion m/z 317 was observed (Fig. 2). As expected, the exit of two sequential CO2 units from both carboxylic acid moieties occurs to give rise the product ions m/z 258 and m/z 214. The last loss of CO2 represents the base peak ion which can undergo steps a, b, and c. Step a is highlighted and provides the neutral elimination of the remaining lateral substituent chain, diagnostic for (3) from the chemical structure. Further loss of an amino unit is predicted in b, while step c presents the detection of m/z 199 which is afforded by another methyl radical loss prior to an internal atom transfer.
MAAs induction using UVRSeveral studies have reported that MAAs biosynthesis in cyanobacteria is induced by PAR and UVR (Rastogi and Incharoensakdi 2014, 2015, Rastogi et al. 2016). When an extract was made from S. torques-reginae ITEP-024 cells exposed to UVR for 3 days, analysis by LC-MS/MS (Geraldes et al. 2020) gave a extracted ion chromatogram typical to that shown in Fig. 3.
The chromatogram showed a significant increase in peak intensities that corresponded to compounds (1), (2), and (3). After UVR induction the concentrations of these MAAs increased substantially from 0.90 to 6.14 μg mg−1. MGA was present in S. torques-reginae ITEP-024 as a minor MAA (<10% of MAA yield). S. torques-reginae ITEP-024 produced MAAs even when not exposed to UVR. This observation suggested constitutive MAA biosynthesis in a quantitative level which has not been previously reported in the literature and might be a natural adaptation by ITEP-024 to extreme UVR-irradiance conditions in Northeastern Brazil where this strain was isolated (Molica et al. 2005, Bassous and Calili 2017). Further experimentation using ITEP-024 are now warranted to determine how environmental and molecular mechanisms converge to induce and then regulate MAA biosynthesis, which currently remain elusive.
Genome annotations and protein predictionsMultiple lines of evidence support that MAAs are derived from conversion of the shikimic acid pathway (Shick and Dunlap 2002, Portwich and Garcia-Pichel 2003, Singh et al. 2008b, Pope et al. 2015). However, contrasting evidence suggests that in cyanobacteria, the parent core structure of mycosporines (4-deoxygadusol) is derived from conversion of the pentose phosphate pathway intermediate sedoheptulose-7-phosphate by the enzymes 2-epi-5-epivaliolone synthase (EVS) and an O-methyltransferase (Balskus and Walsh 2010, Spence et al. 2012, Pope et al. 2015, Katoch et al. 2016). A single cluster of four genes designated mysABCD were identified in the S. torques-reginae ITEP-024 draft genome supporting that these gene products could catalyse the biosynthesis of MAAs. Similarities with publicly available sequences are shown in Table 2 and the organization of the biosynthetic gene cluster is illustrated in Supplementary Fig. S12. In addition, all protein sequences were further annotated using the homology and structural prediction HHpred platform in the MPI Bioinformatic Toolkit (Zimmermann et al. 2018). Based on these analyses and following the biosynthetic scheme proposed by Balskus and Walsh (Balskus and Walsh 2010), mysA was predicted to encode EVS, while mysB encodes the enzyme OMT. Together, these gene products catalyse the biosynthesis of 4-deoxygadusol. The product of gene mysC was an ATP-grasp amino acid ligase predicted to catalyses the addition of glycine to the hydroxyl group at position C3 of 4-deoxygadusol yielding mycosporine-glycine. The cluster also contains mysD, that encodes for a protein with high sequence similarity to cyanobacterial alanine ligases to generate the final product, mycosporine-glycine-alanine. To our knowledge, the function of these enzymes has not been experimentally proven. However, their amino acid sequences share 100% similarity to well characterised bacterial ligases (HHperd analysis) (Zimmermann et al. 2018) which catalyse carbon-nitrogen bond addition of two alanine residues during peptidoglycan biosynthesis. Given the sequence homology between mysD and bacterial alanyl-alanine synthetases, we propose that mysD catalyses a somewhat different reaction, with the addition of a single amino acid residue (serine, threonine, or alanine) to the ketone group at position C1 of mycosporine-glycine to yield shinorine, porphyra-334 or mycosporine-glycine-alanine, respectively.
Bioinformatic evidence that MAA biosynthesis proceeds by conversion of the pentose phosphate pathway intermediate sedoheptulose 7-phosphate has gained increasing traction over recent years, with homologous gene clusters found in many MAA producing organisms and, somewhat surprisingly, bioinformatic and biochemical data strongly supporting biosynthesis of 4-deoxygadusol in zebrafish (Osborn et al. 2015). However, none of these studies have provided evidence that MAA can be abolished following deletion of any of the gene in the cluster. The universal requirement of EVS in the biosynthesis of MAAs was challenged by complete in-frame gene deletion of the EVS gene (Ava_3858) in Anabaena variabilis ATCC 29413 which produces only shinorine (Spence et al. 2012, 2013). Upon UV exposure, shinorine was produced at levels nearly equivalent to that of the wild type strain, suggesting alternative pathways for MAA biosynthesis in A. variabilis. Subsequently, by using pathway specific inhibitors of the shikimate pathway, an alternative route to MAA biosynthesis has been proposed (Fig. 4) (Pope et al. 2015). Constructing an in-frame deletion and screening for complete segregation of the resultant mutant was beyond the scope of the current project, so we also used a pathway specific inhibitor to determine if the shikimate pathway was an alternative route to MAA biosynthesis in S. torques-reginae ITEP-024.
The extract from S. torques-reginae ITEP-024 cells that had been cultivated for 3 days with exposure to UVR in the presence of 2 mM glyphosate (a known shikimate pathway inhibitor) (Bode et al. 1984, Bentley 1990), showed a reduction in peak intensities corresponding to compounds (1), (2), and (3) compared to the control culture where the growth medium had not been supplemented with glyphosate (Supplementary Fig. S13). Hence, using a pathway-specific inhibitor, we demonstrated that MAA biosynthesis might proceed in S. torques-reginae ITEP-024 via the shikimate pathway, supporting a previous study whereby the use of glyphosate abolished MAA biosynthesis in the wild type strain and EVS deletion mutant of the cyanobacterium Anabaena variabilis ATCC 29413 (Pope et al. 2015). Whether glyphosate might induce hitherto unknown MAA catabolic pathways or could repress the promoters of the MAA-producing gene cluster of the pentose phosphate pathway and thereby impede MAA biosynthesis is not known and warrants experimentation.
CONCLUSIONShinorine, porphyra-334, and MGA were detected in culture extracts of S. torques-reginae ITEP-024 and the structure of MGA was fully elucidated. Levels of MAAs could be enhanced after UV irradiance of S. torques-reginae ITEP-024 cultures in agreement with the role of MAAs as natural photo-protective sunscreens. A gene cluster encoded by the genome of S. torques-reginae ITEP-024 suggested involvement of the pentose phosphate pathway in MAA biosynthesis. However, addition of glyphosate inhibited all MAA biosynthesis, supporting a convergent role for the shikimic acid pathway in the MAAs biosynthesis in S. torques-reginae ITEP-024.
ACKNOWLEDGEMENTSThis work was financially supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico-CNPq (311048/2016-1, 141253/2018-5, and 439065/2018-6), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior-CAPES (Grant #23038.001401/2018-92), Fundação de Amparo à Pesquisa do Estado de São Paulo-FAPESP (2018/01563-2, 2015/22742-4 and 2014/50420-9) and University of São Paulo Foundation-FUSP (Project #1979).
SUPPLEMENTARY MATERIALS
Supplementary Fig. S1. Structure of mycosporine-like amino acids isolated from Sphaerospermopsis torques-reginae ITEP-024. 1, shinorine; 2, porphyra-334; 3, mycosporine-glycine-alanine (https://www.e-algae.org).
Supplementary Fig. S2. High performance liquid chromatography-diode array detection chromatogram at 330 nm and UV spectra of a mycosporine-like amino acids extract of Sphaerospermopsis torques-reginae ITEP-024. 1, shinorine; 2, porphyra-334; 3, mycosporine-glycine-alanine (https://www.e-algae.org).
Supplementary Fig. S3. Extracted ion chromatograms (positive electrospray ionization) and UV spectra of target mycosporine-like amino acids. 1, shinorine C13H20N2O8 (0.0 ppm; RT 1.1 min); 2, porphyra-334 C14H22N2O8 (−0.9 ppm; RT 2.2 min); 3, mycosporine-glycine-alanine C13H20N2O7 (0.3 ppm, RT 3.1 min) from Sphaerospermopsis torques-reginae ITEP-024 biomass extract (https://www.e-algae.org).
Supplementary Fig. S4. MS spectra (positive electrospray ionization) of target mycosporine-like amino acids. 1, shinorine C13H20N2O8; 2, porphyra-334 C14H22N2O8; 3, mycosporine-glycine-alanine C13H20N2O7 from Sphaerospermopsis torques-reginae ITEP-024 biomass extract, 35.3 eV (https://www.e-algae.org).
Supplementary Fig. S5. MS/MS spectra (positive electrospray ionization) of target mycosporine-like amino acids. 1, shinorine C13H20N2O8; 2, porphyra-334 C14H22N2O8; 3, mycosporine-glycine-alanine C13H20N2O7 from Sphaerospermopsis torques-reginae ITEP-024 biomass extract, 35.3 eV (https://www.e-algae.org).
Supplementary Fig. S6. 1H NMR spectrum of mycosporine-glycine-alanine in CD3OD (500 MHz) (https://www.e-algae.org).
Supplementary Fig. S7. COSY spectrum of mycosporine-glycine-alanine in CD3OD (500 MHz) (https://www.e-algae.org).
Supplementary Fig. S8. Magnified region of COSY spectrum of mycosporine-glycine-alanine in CD3OD (500 MHz) (https://www.e-algae.org).
Supplementary Fig. S9. Heteronuclear single-quantum correlation spectrum of mycosporine-glycine-alanine in CD3OD (500 MHz) (https://www.e-algae.org).
Supplementary Fig. S10. Magnified region of heteronuclear single-quantum correlation spectrum of mycosporine-glycine-alanine in CD3OD (500 MHz) (https://www.e-algae.org).
Supplementary Fig. S11. Heteronuclear multiple-bond correlation spectrum of mycosporine-glycine-alanine in CD3OD (500 MHz) (https://www.e-algae.org).
Supplementary Fig. S12. Organization of the mycosporine-like amino acids biosynthetic gene cluster in Sphaerospermopsis torques-reginae ITEP-024 (https://www.e-algae.org).
Supplementary Fig. S13. Extracted ion chromatogram (positive electrospray ionization) of a mycosporine-like amino acids extract of Sphaerospermopsis torques-reginae ITEP-024 after exposure to ultraviolet A + ultraviolet B in the presence (dashed line) or absence (full line) of glyphosate (2 mM). 1, shinorine; 2, porphyra-334; 3, mycosporine-glycine-alanine (https://www.e-algae.org).
Fig. 1Observed COSY couplings (A) and heteronuclear multiple-bond correlation (B) for mycosporine-glycine-alanine. 
Fig. 2Plausible fragmentation mechanisms proposed to explain the product ions detected at high resolution tandem mass spectrometry spectra from mycosporine-glycine-alanine (3). 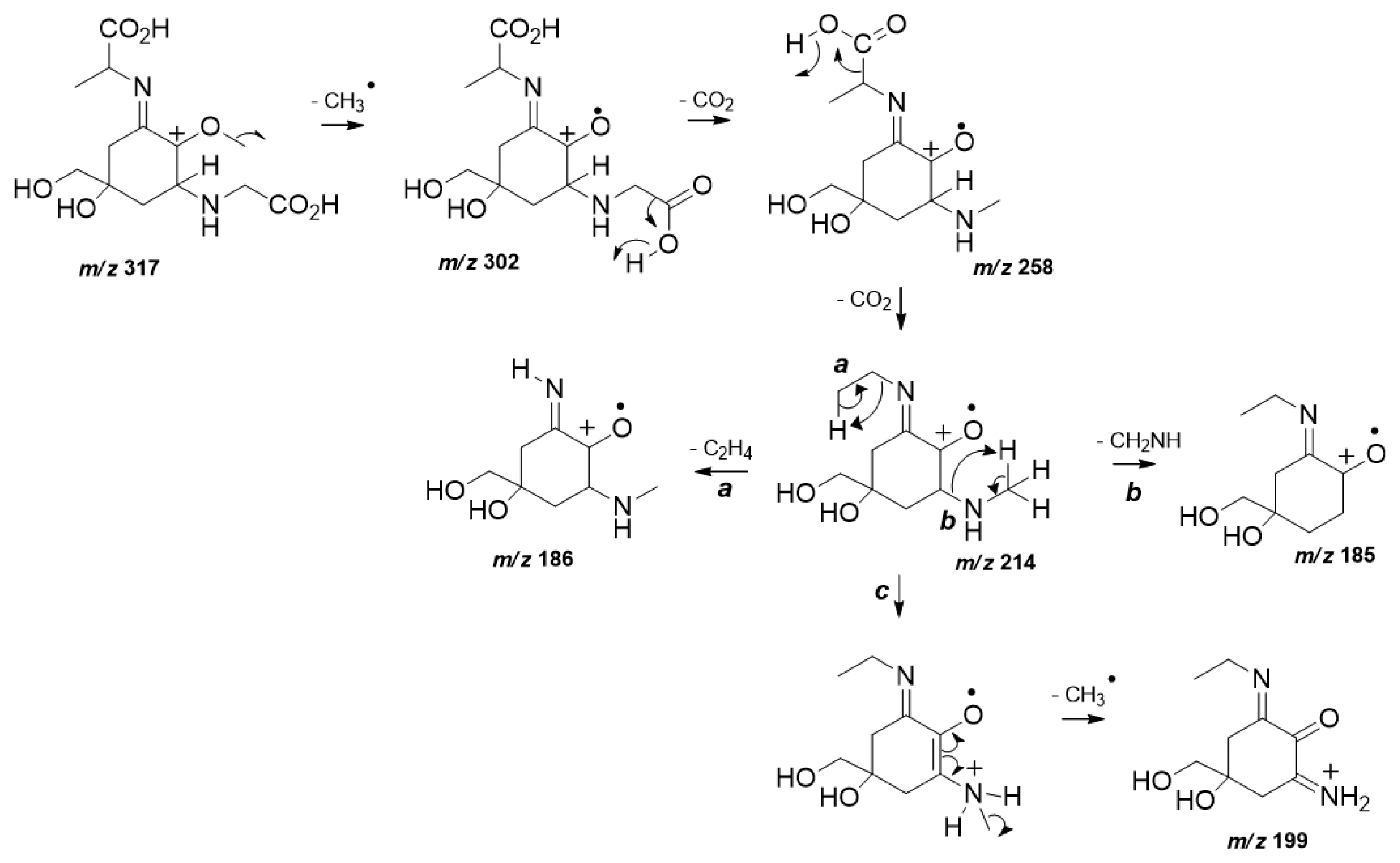
Fig. 3Extracted ion chromatogram (positive electrospray ionization) of a mycosporine-like amino acids extract of Sphaerospermopsis torques-reginae ITEP-024 before exposure (full line) and after exposure (dashed line) to ultraviolet A + ultraviolet B. 1, shinorine; 2, porphyra-334; 3, mycosporine-glycine-alanine. 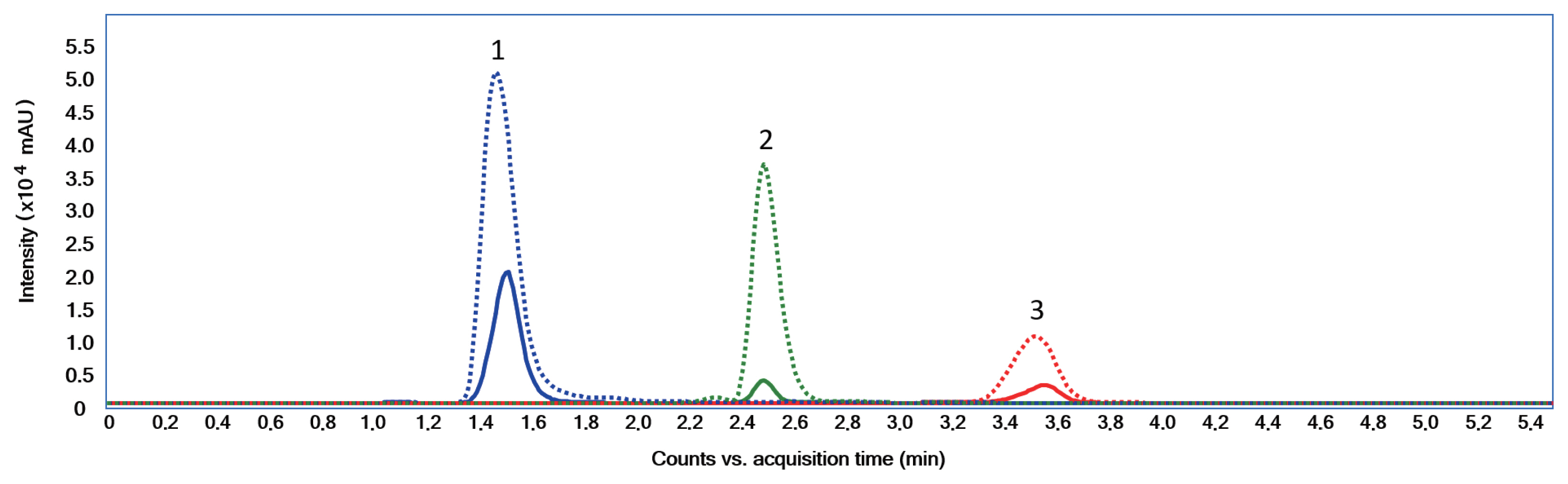
Fig. 4Proposed mycosporine-like amino acids biosynthetic pathway in Sphaerospermopsis torques-reginae ITEP-024. 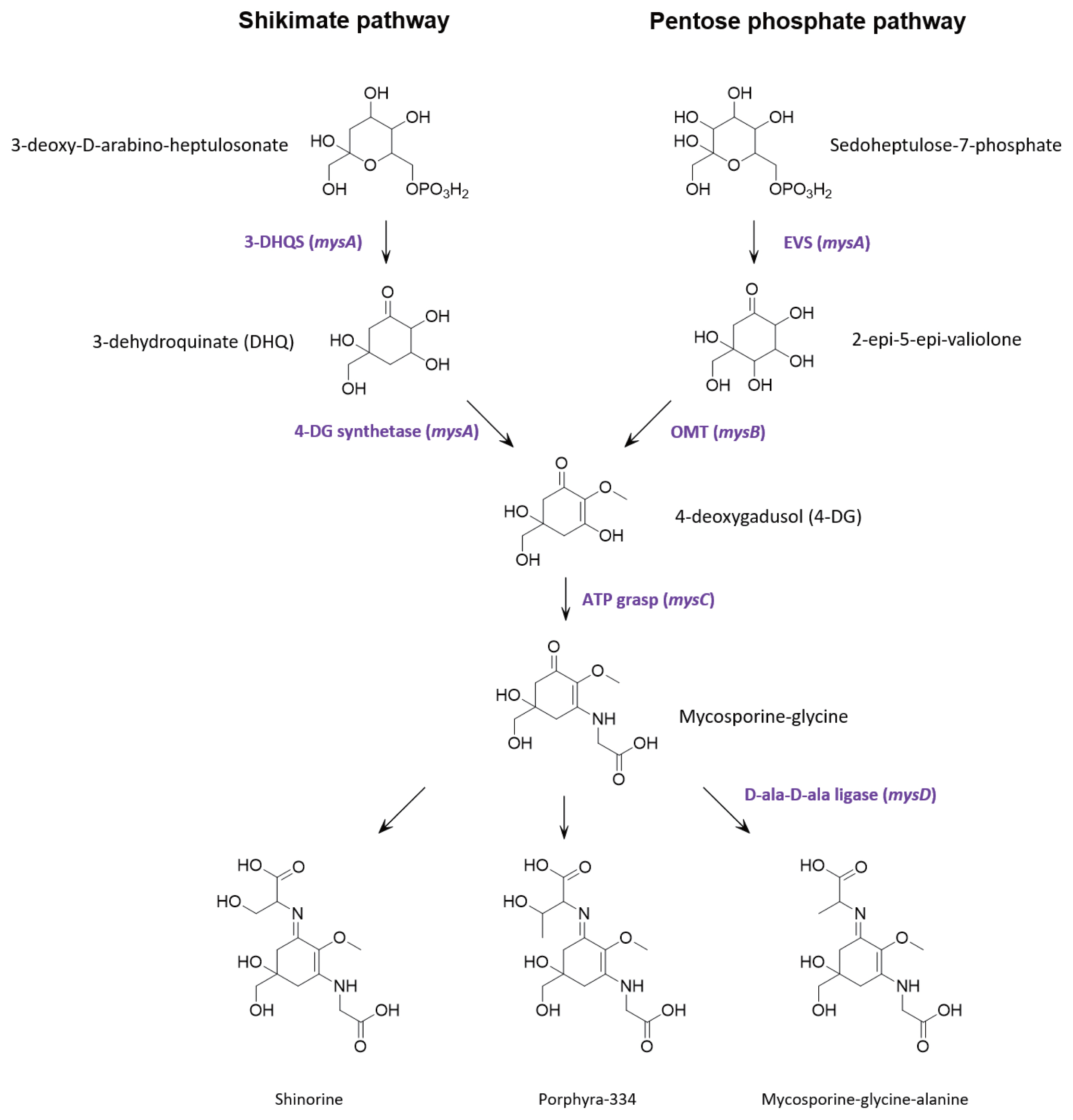
Table 1NMR chemical shifts (500 MHz, CD3OD) of novel MAA
Table 2Proposed function for mysA-D encoded enzymes in the mys biosynthetic gene cluster of Sphaerospermopsis torques-reginae ITEP-024
REFERENCESBalskus, EP. & Walsh, CT. 2010. The genetic and molecular basis for sunscreen biosynthesis in cyanobacteria. Science. 329:1653–1656.
Bassous, GF. & Calili, RF. 2017. Analysis of Brazilian solar irradiance data-characteristic curves of each national grid subsystem. In: New Energy Landscape: Impacts for Latin America, 6th ELAEE/IAEE Latin American Conference, International Association for Energy Economics; Cleveland, OH.
Bentley, R. 1990. The shikimate pathway: a metabolic tree with many branches. Crit Rev Biochem Mol Biol. 25:307–384.
Bode, R., Melo, C. & Birnbaum, D. 1984. Mode of action of glyphosate in Candida maltosa
. Arch Microbiol. 140:83–85.
Boratyn, GM., Camacho, C., Cooper, PS., Coulouris, G., Fong, A., Ma, N., Madden, TL., Matten, WT., McGinnis, SD., Merezhuk, Y., Raytselis, Y., Sayers, EW., Tao, T., Ye, J. & Zaretskaya, I. 2013. BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 41:W29–W33.
Cardozo, KHM., Carvalho, VM., Pionto, E. & Colepicolo, P. 2006. Fragmentation of mycosporine-like amino acids by hydrogen/deuterium exchange and electrospray ionisation tandem mass spectrometry. Rapid Commun Mass Spectrom. 20:253–258.
Cardozo, KHM., Vessecchi, R., Galembeck, SE., Guaratini, T., Gates, PJ., Pinto, E., Lopes, NP. & Colepicolo, P. 2009. A fragmentation study of di-acidic mycosporine-like amino acids in electrospray and nanospray mass spectrometry. J Braz Chem Soc. 20:1625–1631.
Carver, T., Harris, SR., Berriman, M., Parkhill, J. & McQuillan, JA. 2011. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics. 28:464–469.
Cockell, CS. & Knowland, J. 1999. Ultraviolet radiation screening compounds. Biol Rev Camb Philos Soc. 74:311–345.
Dörr, FA., Rodríguez, V., Molica, R., Henriksen, P., Krock, B. & Pinto, E. 2010. Methods for detection of anatoxin-a(s) by liquid chromatography coupled to electrospray ionization-tandem mass spectrometry. Toxicon. 55:92–99.
Geraldes, V., De Medeiros, LS., Jacinavicius, FR., Long, PF. & Pinto, E. 2020. Development and validation of a rapid LC-MS/MS method for the quantification of mycosporines and mycosporine-like amino acids (MAAs) from cyanobacteria. Algal Res. 46:101796 pp.
Geraldes, V., Jacinavicius, FR., Genuàrio, DB. & Pinto, E. 2019. Identification and distribution of mycosporine-like amino acids in Brazilian cyanobacteria using ultrahigh-performance liquid chromatography with diode array detection coupled to quadrupole time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. Advanced online publication.
https://doi.org/10.1002/rcm.8634
Gorham, PR., McLachlan, J., Hammer, UT. & Ki, WK. 1964. Isolation and culture of toxic strains of Anabaena flos-aquae (Lyngb.) de Bréb: with plate 7, 1 figure and 3 tables in the text. Int Ver Theor Angew Limnol Verh. 15:796–804.
Katoch, M., Mazmouz, R., Chau, R., Pearson, LA., Pickford, R. & Neilan, BA. 2016. Heterologous production of cyanobacterial mycosporine-like amino acids mycosporine-ornithine and mycosporine-lysine in Escherichia coli
. Appl Environ Microbiol. 82:6167–6173.
Laatsch, H. 2014. AntiBase 2014: The Natural Compound Identifier, Available from: https://application.wiley-vch.de/stmdata/antibase.php
. Accessed May 1, 2020
Lawrence, KP., Gacesa, R., Long, PF. & Young, AR. 2018. Molecular photoprotection of human keratinocytes in vitro by the naturally occurring mycosporine-like amino acid palythine. Br J Dermatol. 178:1353–1363.
Lawrence, KP., Long, PF. & Young, AR. 2017. Mycosporine-like amino acids for skin photoprotection. Curr Med Chem. 25:5512–5527.
Lima, ST., Alvarenga, DO., Etchegaray, A., Fewer, DP., Jokela, J., Varani, AM., Sanz, M., Dörr, FA., Pinto, E., Sivonen, K. & Fiore, MF. 2017. Genetic organization of anabaenopeptin and spumigin biosynthetic gene clusters in the cyanobacterium Sphaerospermopsis torques-reginae ITEP-024. ACS Chem Biol. 12:769–778.
Miyamoto, KT., Komatsu, M. & Ikeda, H. 2014. Discovery of gene cluster for mycosporine-like amino acid biosynthesis from Actinomycetales microorganisms and production of a novel mycosporine-like amino acid by heterologous expression. Appl Environ Microbiol. 80:5028–5036.
Molica, RJR., Oliveira, EJA., Carvalho, PVVC., Costa, ANSF., Cunha, MCC., Melo, GL. & Azevedo, SMFO. 2005. Occurrence of saxitoxins and an anatoxin-a(s)-like anticholinesterase in a Brazilian drinking water supply. Harmful Algae. 4:743–753.
Orfanoudaki, M., Hartmann, A., Karsten, U. & Ganzera, M. 2019. Chemical profiling of mycosporine-like amino acids in twenty-three red algal species. J Phycol. 55:393–403.
Osborn, AR., Almabruk, KH., Holzwarth, G., Asamizu, S., LaDu, J., Kean, KM., Karplus, PA., Tanguay, RL., Bakalinsky, AT. & Mahmud, T. 2015. De novo synthesis of a sunscreen compound in vertebrates. eLife. 4:e05919 pp.
Pope, MA., Spence, E., Seralvo, V., Gacesa, R., Heidelberger, S., Weston, AJ., Dunlap, WC., Shick, JM. & Long, PF. 2015. O-Methyltransferase is shared between the pentose phosphate and shikimate pathways and is essential for mycosporine-like amino acid biosynthesis in Anabaena variabilis ATCC 29413. ChemBioChem. 16:320–327.
Portwich, A. & Garcia-Pichel, F. 2003. Biosynthetic pathway of mycosporines (mycosporine-like amino acids) in the cyanobacterium Chlorogloeopsis sp. strain PCC 6912. Phycologia. 42:384–392.
Rastogi, RP. & Incharoensakdi, A. 2014. UV radiation-induced biosynthesis, stability and antioxidant activity of mycosporine-like amino acids (MAAs) in a unicellular cyanobacterium Gloeocapsa sp. CU2556. J Photochem Photobiol B Biol. 130:287–292.
Rastogi, RP. & Incharoensakdi, A. 2015. Occurrence and induction of a ultraviolet-absorbing substance in the cyanobacterium Fischerella muscicola TISTR8215. Phycol Res. 63:51–55.
Rastogi, RP., Madamwar, D., Nakamoto, H. & Incharoensakdi, A. 2019. Resilience and self-regulation processes of microalgae under UV radiation stress. J Photochem Photobiol C Photochem Rev. Advanced online publication.
https://doi.org/10.1016/j.jphotochemrev.2019.100322
Rastogi, RP., Richa, ., Kumar, A., Tyagi, MB. & Sinha, RP. 2010. Molecular mechanisms of ultraviolet radiation-induced DNA damage and repair. J Nucleic Acids. 2010:592980.
Rastogi, RP., Sinha, RP., Moh, SH., Lee, TK., Kottuparambil, S., Kim, Y., Rhee, J., Choi, E., Brown, MT., Häder, D. & Han, T. 2014. Ultraviolet radiation and cyanobacteria. J Photochem Photobiol B Biol. 141:154–169.
Rastogi, RP., Sonani, RR., Mandamwa, D. & Incharoensakdi, A. 2016. Characterization and antioxidant functions of mycosporine-like amino acids in the cyanobacterium Nostoc sp. R76DM. Algal Res. 16:110–118.
Sanz, M., Dörr, FA. & Pinto, E. 2015. First report of spumigin production by the toxic Sphaerospermopsis torques-reginae cyanobacterium. Toxicon. 108:15–18.
Sanz, M., Salinas, RK. & Pinto, E. 2017. Namalides B and C and spumigins K-N from the cultured freshwater cyanobacterium Sphaerospermopsis torques-reginae
. J Nat Prod. 80:2492–2501.
Sayers, EW., Agarwala, R., Bolton, EE., Brister, JR., Canese, K., Clark, K., Connor, R., Fiorini, N., Funk, K., Hefferon, T., Holmes, JB., Kim, S., Kimchi, A., Kitts, PA., Lathrop, S., Lu, Z., Madden, TL., Marchler-Bauer, A., Phan, L., Schneider, VA., Schoch, CL., Pruitt, KD. & Ostell, J. 2019. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 47:D23–D28.
Shick, JM. & Dunlap, WC. 2002. Mycosporine-like amino acids and related gadusols: biosynthesis, accumulation, and UV-protective functions in aquatic organisms. Annu Rev Physiol. 64:223–262.
Shick, JM., Romaine-Lioud, S., Romaine-Lioud, S., Ferrier-Pagès, C. & Gattuso, J-P. 1999. Ultraviolet-B radiation stimulates shikimate pathway-dependent accumulation of mycosporine-like amino acids in the coral Stylophora pistillata despite decreases in its population of symbiotic dinoflagellates. Limnol Oceanogr. 44:1667–1682.
Singh, SP., Klisch, M., Sinha, RP. & Häder, D. 2008a. Effects of abiotic stressors on synthesis of the mycosporine-like amino acid shinorine in the cyanobacterium Anabaena variabilis PCC 7937. Photochem Photobiol. 84:1500–1505.
Singh, SP., Kumari, S., Rastogi, RP., Sinha, KL. & Sinha, R. 2008b. Mycosporine-like amino acids (MAAs): chemical structure, biosynthesis and significance as UV-absorbing/screening compounds. Indian J Exp Biol. 46:7–17.
Spence, E., Bryan, SJ., Lisfi, M., Cullum, J., Dunlap, WC., Shick, JM., Mullineaux, CW. & Long, PF. 2013. 2-epi-5-epi-Valiolone synthase activity is essential for maintaining phycobilisome composition in the cyanobacterium Anabaena variabilis ATCC 29413 when grown in the presence of a carbon source. Photosynth Res. 116:33–43.
Spence, E., Dunlap, WC., Shick, JM. & Long, PF. 2012. Redundant pathways of sunscreen biosynthesis in a cyanobacterium. Chembiochem. 13:531–533.
Taylor & Francis Group 2017. Dictionary of Natural Products 28.1, CRC Press, Available from: http://dnp.chemnetbase.com/faces/chemical/ChemicalSearch.xhtml
. Accessed May 1, 2020
Werner, VR., Laughinghouse, HD. 4th, Fiore, MF., Sant’Anna, CL., Hoff, C., de Souza Santos, KR., Neuhaus, EB., Molica, RJR., Honda, RY. & Echenique, RO. 2012. Morphological and molecular studies of Sphaerospermopsis torques-reginae (Cyanobacteria, Nostocales) from South American water blooms. Phycologia. 51:228–238.
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||