INTRODUCTION
The genus Mayamaea was proposed by Lange-Bertalot (1997). Initially, 13 taxa formerly attributed to Navicula sensu lato were included in the genus (Lange-Bertalot 1997). Mayamaea includes small-celled species that are elliptical in shape. The main characteristic of the genus is the arrangement of the pore occlusions, which are hymenes that cover the areolae from the outside, as well as the presence of a pronounced sternum found in the vast majority of species. The location of the hymenes is an important characteristic that differentiates Mayamaea from the genera Eolimna Lange-Bertalot and W. Schiller and Sellaphora Mereschkowsky, otherwise these genera have much in common including the structure of the raphe, central pores and distal raphe ends (Barragán et al. 2017). Therefore, accurate identification by morphological characteristics is possible only when studying the material using a scanning electron microscope. Currently, the genus has about 30 taxa (Guiry and Guiry 2020, Kociolek et al. 2020). Species of the genus Mayamaea are distributed worldwide, though most species are known from Europe (Barragán et al. 2017). The species Mayamaea agrestis (Hustedt) Lange-Bertalot (basionym: Navicula agrestis Hustedt) was described from Southeast Asia (Indonesia) (Hustedt in Schmidt 1936, Hustedt 1937).
Species of Mayamaea prefer humid terrestrial habitats, as well as freshwater ecosystems (oligotrophic and mesotrophic), including in the littoral zone of these environments (Lange-Bertalot 1997, Kulikovskiy 2006, Kulikovskiy et al. 2016). In discussing the ecology of the species, Barragán et al. (2017) suggest that soil habitats are typical for M. lacunolaciniata (Lange-Bertalot and Bonik) Lange-Bertalot, M. muraliformis Lange-Bertalot, M. terrestris N. Abarca and R. Jahn, and M. petersenii Barragán, Ector and C. E. Wetzel. However, some other species have also been found in the soil. For example, M. atomus (Kutzing) Lange-Bertalot was found in the soil of paddy fields in Central Japan (Ohtsuka and Fujita 2001). A large number of diatom species (147 taxa) were identified from cultivated soils in Podkarpacie Province, Poland (Noga et al. 2014), and the most numerous was M. atomus and 5 other species. Others species of Mayamaea were found as well, including M. agrestis (Hustedt) Lange-Bertalot, M. excelsa (Krasske) Lange-Bertalot, M. permitis (Hustedt) Bruder & Medlin, M. cf. recondita (Hustedt) Lange-Bertalot, Mayamaea sp 2. Foets et al. (2020) found M. atomus (Kützing) Lange-Bertalot as a dominant in the soil of the Attert River basin (Luxembourg). The authors revealed that M. agrestis (Hustedt) Lange-Bertalot was typically on agricultural grasslands, M. permitis (Hustedt) Bruder & Medlin was one of the most abundant species in the territory, and M. fossalis (Krasske) was found in undisturbed grassland soils.
Molecular genetic data on Mayamaea species are scarce. A study of two strains of M. atomus (Kützing) Lange-Bertalot and one strain of M. atomus var. permitis (Hustedt) Lange-Bertalot showed that they form a monophyletic group, separate from but related to representatives of Pinnularia Ehrenberg and Caloneis Cleve (Bruder and Medlin 2008). Further, the results of studying the molecular markers 18S V4 and rbcL for strains of M. atomus, M. permitis (Hustedt) Bruder and Medlin, M. fossalis (Krasske) Lange-Bertalot, and M. terrestris were summarized by Zimmermann et al. (2014). The authors included their own sequences and all available data from the INSDC database. It was shown that all strains identified as M. terrestris and M. permitis form an independent clade between Navicula Bory and Caloneis. Currently, in the NCBI database 59 sequences for seven taxa have been deposited: M. atomus, M. permitis, M. fossalis, M. terrestris, Mayamaea sp. KAS1111, Mayamaea sp. DM-MEX020, and Mayamaea sp. DM-MEX019.
We also note that the only mention of soil diatoms from Southeast Asia (Indonesia) refers to the work of Kolkwitz and Krieger (1936), where the authors used one soil sample (“Humus unter Bäumen”, Humus under the trees, 2,700 m, pH = 4.3) from Mount Pangrango. The following species were identified by the authors: Eunotia pectinalis (Kützing) Rabenhorst, E. praerupta var. thermalis Hustedt, Melosira dickiei (Thwaites) Kützing, Navicula contenta Grunow in Van Heurck and Pinnularia lata var. thuringiaca (Rabenhorst) Ant. Mayer.
Previously, we described species of the genera Aulacoseira Thwaites, Eunotia Ehrenberg, Gomphonema Ehrenberg and Luticola D. G. Mann (Glushchenko et al. 2016, 2017a, 2017b, 2018) from freshwater reservoirs of Cát Tiên National Park (Vietnam). However, the present report is the first study on the soil algae in this area.
This work provides a morphological description and molecular data on the 18S V4 and rbcL genetic markers for a new species from the genus Mayamaea isolated into a monoclonal culture from a soil sample from the territory of Cát Tiên National Park. The work is the first, and part of a larger project, on the study of soil algae in Vietnam. To date, 57 soil samples from the national park have been studied and 565 algae strains have been isolated.
MATERIALS AND METHODS
Study area
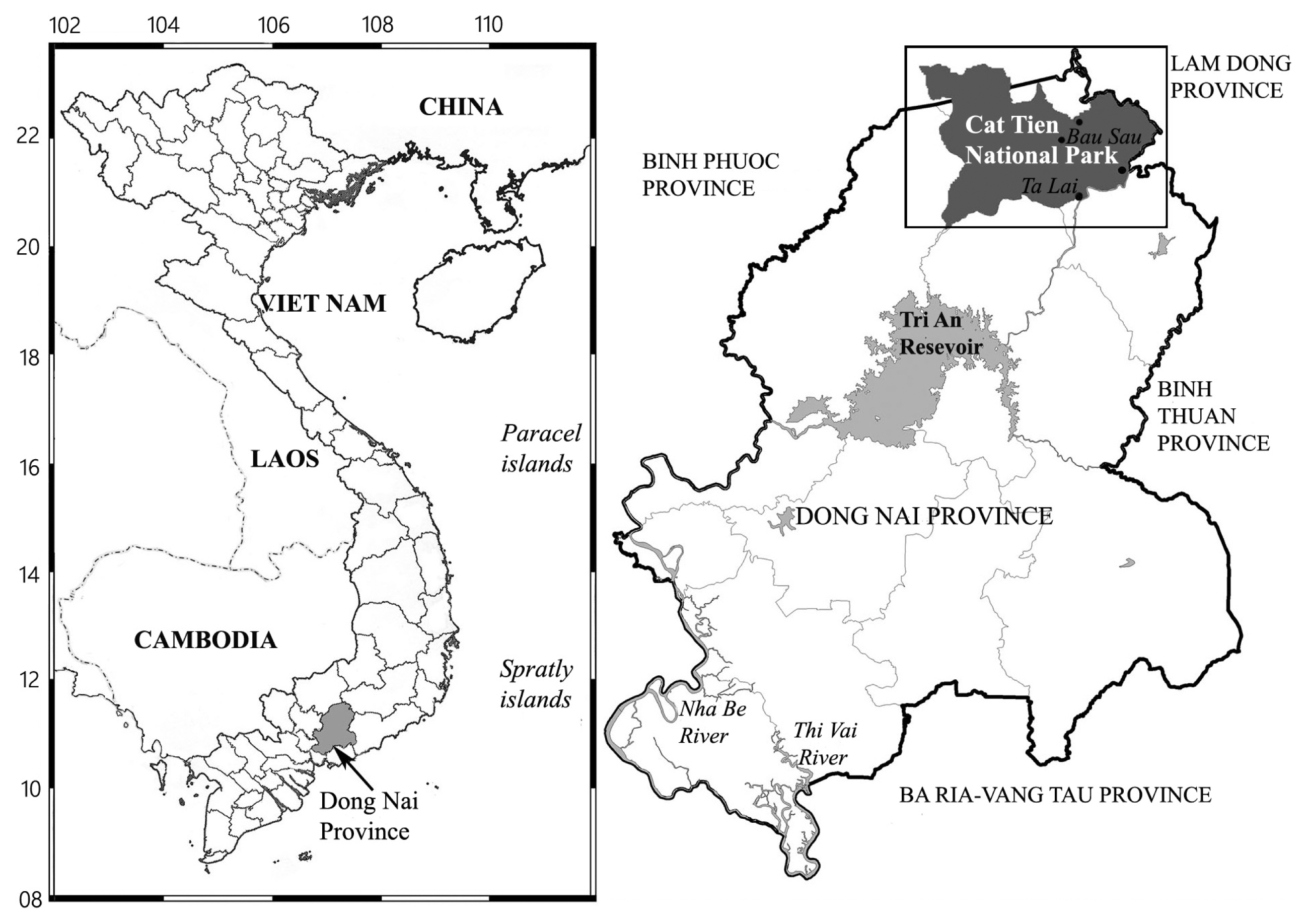
Cát Tiên National Park is located in southern Vietnam, 150 km northeast of Ho Chi Minh City (Fig. 1). The park was established in 1978 and consists of three parts with a total area of 73,878 ha (Blanc et al. 2001). The region belongs to the bioclimatic type of monsoon tropical climate with summer rains. Relative humidity, as a rule, always exceeds 70%, with an average annual temperature of about 26°C. From December to March there is almost no rainfall. Southwest monsoons last eight months, from April to November, when most of the precipitation is observed. The wet season peaks in August-September. At this time of the year, up to 400–450 mm of precipitation falls per month, which leads to flooding of a significant part of the park. The main part of the territory is occupied by forests, which are of the monsoon, semi-deciduous type. These forests are characterized by high biological diversity and high productivity, second only to moist tropical forests in this respect (Blanc et al. 2001).
Sampling
The sample used in the present report was collected from Vietnam by E. S. Gusev, Jun 9, 2018, and designated No. 06460, from the Cát Tiên National Park, Đồng Nai Province (11°28.820′ N, 107°22.836′ E). It was comprised of wet soil, which had a pH of 6.5. Samples in general were taken as follows: first, the surface of the test site was examined in order to detect macrogrowth of algae, then a combined sample was taken from an area of 10–30 m2 using the metal scoop or shovel. The composite sample consists of 5–10 individual samples. For an individual sample, the topsoil was removed from an area of 5 to 20 cm2. After selection, the instruments were cleaned and sterilized with ethanol. Samples were placed in plastic zip bags, labeled. Immediately after the selection, the absolute humidity was determined in the laboratory room by the “hot drying” method (Vadjunina and Korchagina 1986), then air dried and packaged.
Measurement of pH
To measure pH, we weighed 30 g of soil to which 150 mL of distilled water was added. The suspension was poured into a clean glass and measurements were made using the Hanna Combo device (HI 98129; Hanna Instruments, Inc., Woonsocket, RI, USA) (Arinushkina 1970).
Culturing
Isolation and cultivation of algae strains was carried out at the Institute of Plant Physiology Russian Academy of Sciences. A subsample of each collection was added to WC liquid medium (Guillard and Lorenzen 1972). Monoclonal strains were established by micropipetting single cells under an inverted microscope. Non-axenic unialgal cultures were maintained in WC liquid medium at 22–25°C in a growth chamber with a 12 : 12 h light : dark photoperiod.
Preparation of slides and microscope investigation
The culture was treated with 10% hydrochloric acid to remove carbonates and washed several times with deionized water for 12 h. Afterwards, the sample was boiled in concentrated hydrogen peroxide (≈37%) to remove organic matter. It was washed again with deionized water four times at 12 h intervals. After decanting and filling with deionized water up to 100 mL, the suspension was pipetted onto coverslips and left to dry at room temperature. Permanent diatom preparations were mounted in Naphrax. Light microscopic (LM) observations were performed with a Zeiss Axio Scope A1 microscope (Carl Zeiss Microscopy GmbH, Gottingen, Germany) equipped with an oil immersion objective (×100, n.a. 1.4, differential interference contrast) and Axiocam ERc 5s camera (Carl Zeiss Microscopy GmbH). Valve ultrastructure was examined using a scanning electron microscope JSM-6510LV (IBIW; Institute for Biology of Inland Waters RAS, Borok, Russia). For scanning electron microscopy (SEM), part of the suspensions was fixed on aluminum stubs after air-drying. The stubs were sputter-coated with 50 nm of Au using an Eiko IB 3 machine (Eiko Engineering Co. Ltd., Tokyo, Japan).
The suspension and slides are deposited in the collection of Maxim Kulikovskiy at the Herbarium of the Institute of Plant Physiology Russian Academy of Sciences, Moscow, Russia.
Extraction of DNA and amplification
Total DNA of monoclonal cultures was extracted using InstaGene Matrix (Bio-Rad, Hercules, CA, USA) according to the manufacturer’s protocol. Fragments of 18S rDNA (323 bp, including V4 domain) and partial rbcL plastid genes (978 bp) were amplified using primers D512for and D978rev from Zimmerman et al. (2011) for 18S rDNA fragments and rbcL40+ from Ruck and Theriot (2011) and rbcL1255- from Alverson et al. (2007) for rbcL fragments.
Amplifications of the 18S rDNA fragments and partial rbcL gene fragment were carried out using the premade mix ScreenMix (Evrogen, Moscow, Russia) for the polymerase chain reaction (PCR). The conditions of amplification for 18S rDNA fragments were: an initial denaturation of 5 min at 95°C, followed by 35 cycles at 94°C for denaturation (30 s), 52°C for annealing (30 s) and 72°C for extension (50 s), and a final extension of 10 min at 72°C. The conditions of amplification for partial rbcL were: an initial denaturation of 5 min at 95°C, followed by 45 cycles at 94°C for denaturation (30 s), 59°C for annealing (30 s) and 72°C for extension (80 s), and a final extension of 10 min at 72°C.
The resulting amplicons were visualized by horizontal agarose gel electrophoresis (1.5%), colored with SYBR Safe (Life Technologies, Carlsbad, CA, USA). Purification of DNA fragments was performed with the ExoSAP-IT kit (Affimetrix, Santa Clara, CA, USA) according to the manufacturer’s protocol. 18S rDNA fragments and partial rbcL gene were decoded from two sides using forward and reverse PCR primers and the Big Dye system followed by electrophoresis using a Genetic Analyzer 3500 sequencer (Applied Biosystems, Foster City, CA, USA).
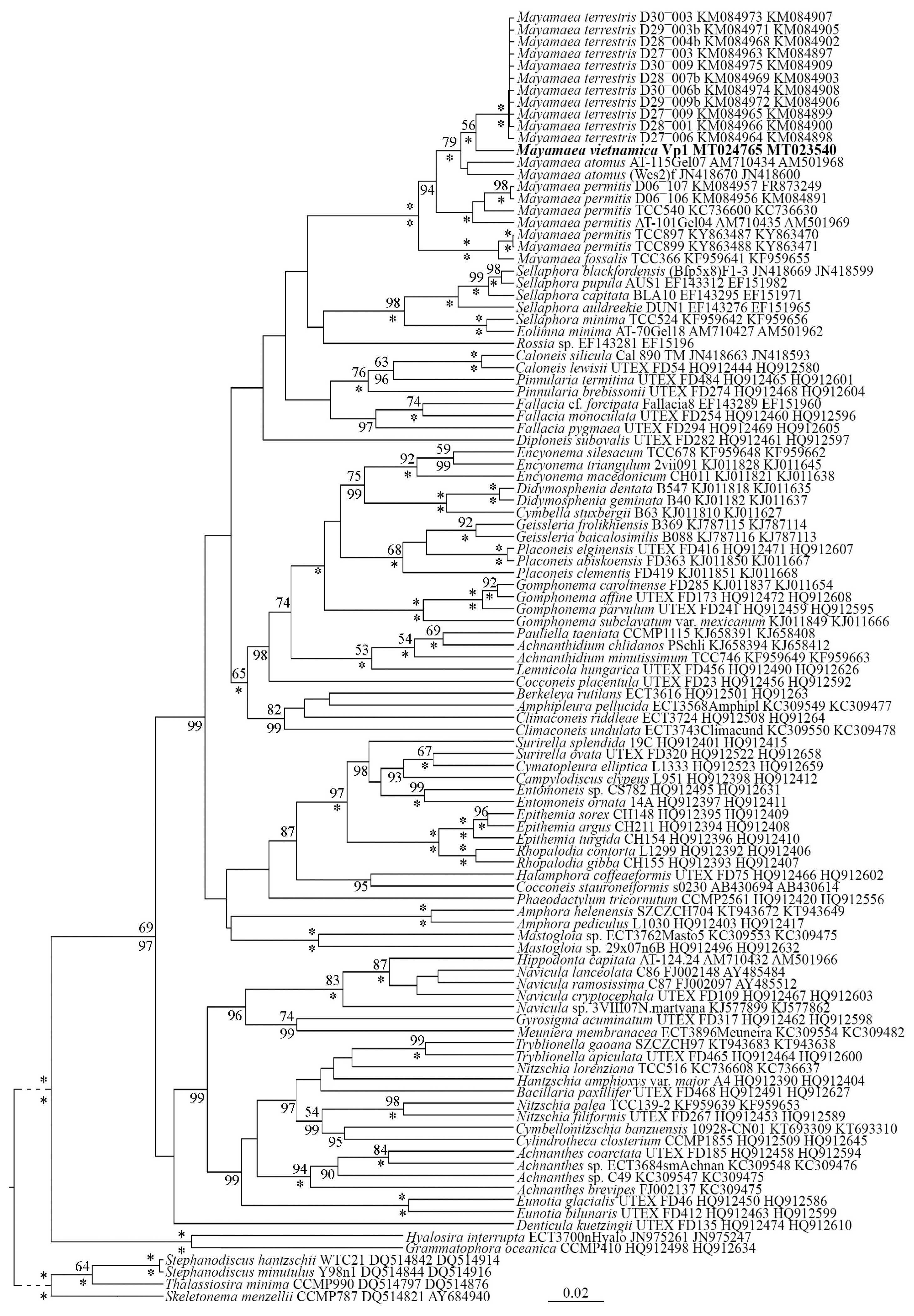
Editing and assembling of the consensus sequences were carried out by comparing the direct and reverse chromatograms using the Ridom TraceEdit program ver. 1.1.0 (Ridom GmbH, Würzburg, Germany) and Mega7 (Kumar et al. 2016). Newly determined sequences and DNA fragments of 106 other diatoms, which were downloaded from GenBank (taxa and accession numbers are given in the tree) (Fig. 2), were included in the alignments. Four centric diatom species were chosen as the outgroups.
The nucleotide sequences of the 18S rDNA and rbcL genes were aligned separately using the Mafft v7 software and the E-INS-i model (Katoh and Toh 2010). For the protein-coding sequences of the rbcL gene, we checked that the beginning of the aligned matrix corresponded to the first position of the codon (triplet). The resulting alignments had lengths of 402 (18S rDNA) and 978 (rbcL) characters.
The data set was analyzed using Bayesian inference (BI) method implemented in Beast ver. 1.10.1. (Drummond and Rambaut 2007) to construct the phylogeny. For each of the alignment partitions, the most appropriate substitution model was estimated using the Bayesian information criterion (BIC) as implemented in jModelTest 2.1.10 (Darriba et al. 2012). This BIC-based model selection procedure selected the following models, shape parameter α and a proportion of invariable sites (pinvar): TrN + I + G, α = 0.4410 and pinvar = 0.3730 for 18S rDNA; TIM1 + I + G, α = 0.5030 and pinvar = 0.5960 for the first codon position of the rbcL gene; TIM1ef + I + G, α = 0.2490 and pinvar = 0.6820 for the second codon position of the rbcL gene; TVM + I + G, α = 0.7500 and pinvar = 0.1150 for the third codon position of the rbcL gene. We used the GTR model of nucleotide substitution instead of TIM1, TIM1ef and TVM, given that they were the best matching model available for BI method. A Yule process tree prior was used as a speciation model. The analysis ran for 10 million generations with chain sampling every 1,000 generations. The parameters-estimated convergence, effective sample size and burn-in period were checked using the software Tracer ver. 1.7.1. (Drummond and Rambaut 2007). The initial 25% of the trees were removed, the rest retained to reconstruct a final phylogeny. The phylogenetic tree and posterior probabilities of its branching were obtained on the basis of the remaining trees, having stable estimates of the parameter models of nucleotide substitutions and likelihood. Maximum likelihood (ML) analysis was performed using the program RAxML (Stamatakis et al. 2008). The nonparametric bootstrap analysis with 1,000 replicas was used. The statistical support values were visualized in FigTree ver. 1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/) and Adobe Photoshop CC 19.0 (Adobe Systems, San José, CA, USA).
RESULTS
Mayamaea vietnamica Glushchenko, Kezlya, Kulikovskiy & Kociolek sp. nov. (Figs 3–5)
Holotype
Collection of Maxim Kulikovskiy at the Herbarium of the Institute of Plant Physiology Russian Academy of Science, Moscow, Russia, holotype here designated, slide No. 06460 = Fig. 3K. Strain VP 1 isolated from forest soil.
Type locality
Vietnam, Cát Tiên National Park, Đồng Nai Province, forest soil (11°28.820′ N, 107°22.836′ E). Leg. E. S. Gusev, Jun 9, 2018.
Distribution
As yet known only from the type locality.
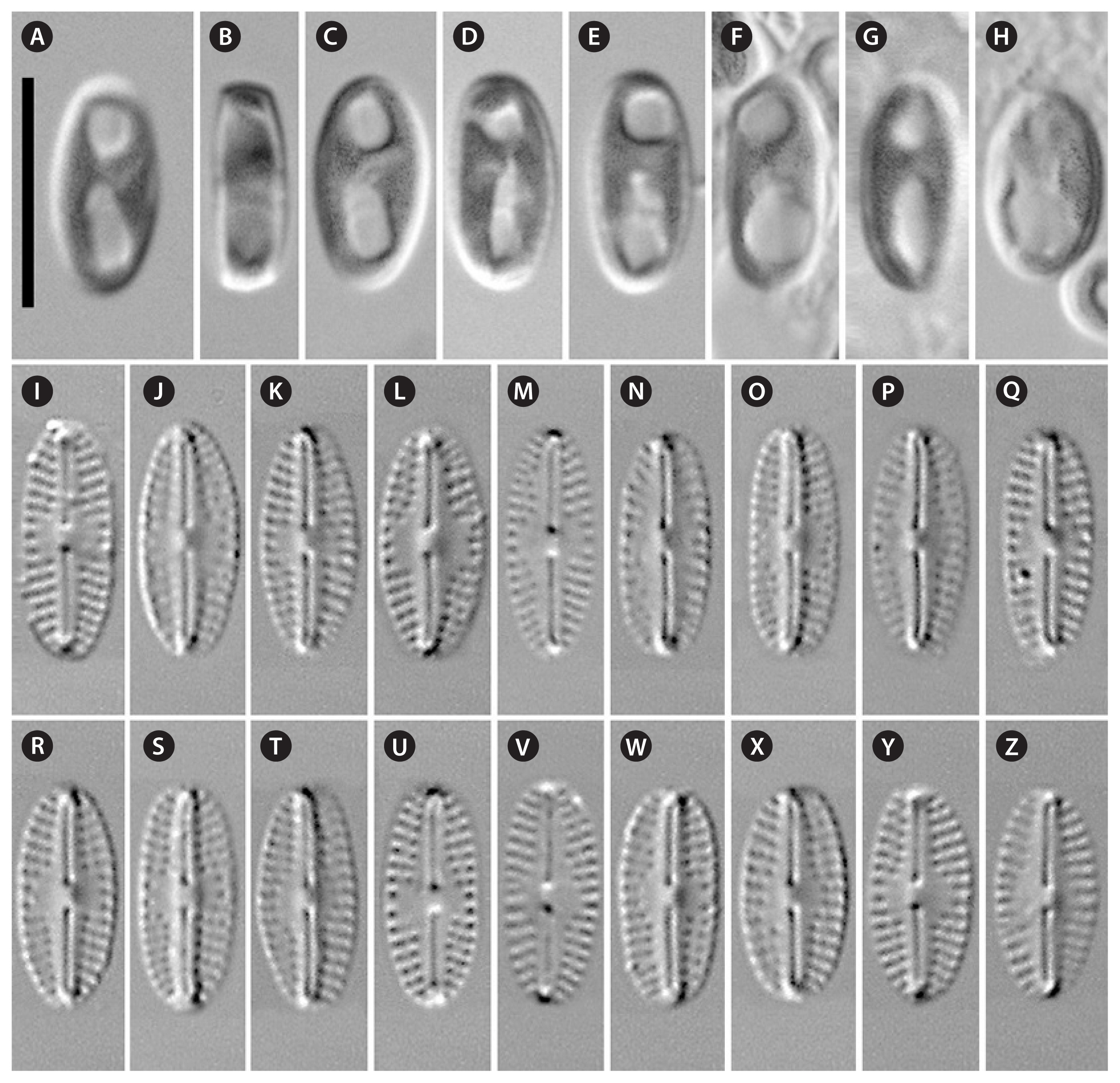
Description. LM (Fig. 3A–Z)
Cells solitary, rectangular in girdle view (Fig. 3B). One H-shaped plastid, with one arm lying against each side of the girdle, connected by a narrow central isthmus (Fig. 3A & C–H). Valves small, from almost linear to elliptical and oval (n = 29) with pronounced sternum and broadly rounded ends. Length 9.1–10.5 μm, breadth 3.9–4.8 μm. Raphe narrow, linear. Central area more or less expressed, rounded to asymmetrical, rarely transversally elongated, and bordered on each margin by 3 shortened striae and / or 3 isolated areolae. Axial area tapers from the central area, becoming narrower towards the ends. The striae are radiate throughout the valve, 19–22 in 10 μm. Areolae coarse, clearly visible in LM.
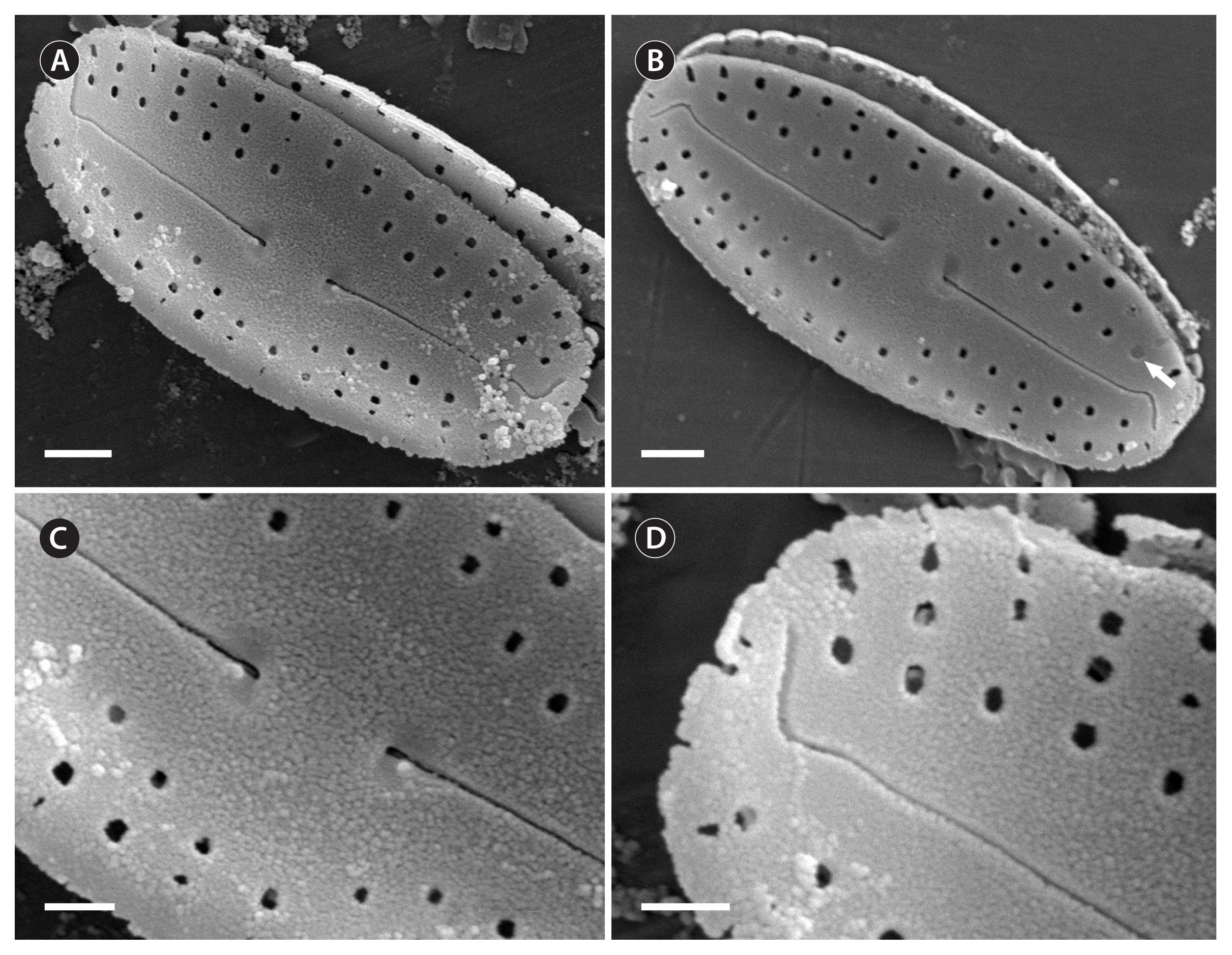
SEM, external views (Fig. 4A–D)
Raphe narrow, linear (Fig. 4A & B). Proximal raphe ends straight, drop-shaped (Fig. 4C). Distal raphe fissures bending strongly to the same side of valve mantle (Fig. 4A, B & D). Striae composed in 1–3 round areolae, not extending to valve margin (Fig. 4A–D). Areolae round, covered by individual hymenes (example, Fig. 4B, white arrow).
SEM, internal views (Fig. 5A–D)
Raphe narrow, linear (Fig. 4A). Proximal valve ends slightly deflected to one side (Fig. 4B). Distal raphe ends terminating in small helictoglossae (Fig. 4C & D).
Molecular investigation (Fig. 2)
Phylogenetic analysis of molecular sequence data shows that our new taxon, M. vietnamica sp. nov., is part of a branch with other species of Mayamaea (ML, 56; BI, 100). The branch with Mayamaea species is well-supported (ML, 100; BI, 100) and independent from other branches. Closely related to the monophyletic group of Mayamaea species is a branch with strains of species from the genera Sellaphora and Eolimna. The branch containing these genera also has high statistical support (ML, 98; BI, 100). Six strains of M. permitis are divided in two branches in our molecular tree. This suggests the presence of at least two cryptic species from specimens whose morphology suggest they could be assigned to the taxon M. permitis sensu lato. M. fossalis is close to two strains of M. permitis, however a difference between these species is supported by high statistical support (ML, 100; BI, 100). This result shows that these two species are independent taxa based on this current molecular investigation.
DISCUSSION
Mayamaea vietnamica sp. nov. possesses all of the typical characters of the genus Mayamaea (Lange-Bertalot 1997). These features include hymenes that covers the areolae from the outside, uniseriate striae, large areolae, small size, as well as the presence of a pronounced sternum. Our new species is morphologically similar to some known species from the genus Mayamaea, but it differs from them by both quantitative and qualitative features (see Table 1). The results of our molecular investigation show the new species is part of an independent branch the includes three taxa. M. vietnamica sp. nov. is more closely related to M. terrestris than either is to M. atomus. Additionally, M. permitis and M. fossalis were included in the molecular investigation. The lineage of Mayamaea taxa is part of a broader monophyletic group that includes species of the genera Sellaphora, Eolimna and Rossia M. Voigt (see Fig. 2). However, the results of our molecular investigation show that there is high statistical support for recognizing Mayamaea as a genus distinct from these other genera (see above).
In Table 1, we compare species of Mayamaea that are morphologically similar to our new species described herein. The type species of Mayamaea is M. atomus and this species is similar in valve shape to M. vietnamica sp. nov. However, M. vietnamica sp. nov. differs from M. atomus in general by having a lower density of areolae (20–30 at 10 μm in M. vietnamica sp. nov. vs. 40–50 at 10 μm in M. atomus), as well as a lower number of areolae in the striae (1–3 in M. vietnamica sp. nov. vs. 1–8 in M. atomus). In addition, the axial area of M. vietnamica sp. nov. is much wider than that of M. atomus, whose axial area is narrow, corresponds to the sternum, while in M. vietnamica sp. nov. the axial area is markedly expressed, tapering from the central area becoming narrower towards the ends of the valves. The central area of M. atomus is small or absent, which is not observed in M. vietnamica sp. nov., the central area of which is always noticeably pronounced.
The species Mayamaea crassistriata Lange-Bertalot, Cavacini, Tagliaventi and Alfinito (Lange-Bertalot et al. 2003) is similar to M. vietnamica sp. nov. and both taxa have axial areas that broaden into to the central area. However, a central area is absent in M. crassistriata, while in M. vietnamica sp. nov. it is well expressed. In addition, the axial area is much wider in M. vietnamica sp. nov. M. crassistriata is characterized by a higher density of areolae (ca. 40 in 10 μm vs. 20–30 in 10 μm in M. vietnamica sp. nov.). The number of areolae in striae is also higher in M. crassistriata (which has 2–6 vs. 1–3 in M. vietnamica sp. nov.).
The morphologically most similar species to M. vietnamica sp. nov. is Mayamaea fossalis (Krasske 1929, p. 354, fig. 10; Lange-Bertalot et al. 1996, p. 109, 110, Pl. 19, figs 14 & 15; Lange-Bertalot 2001, p. 138, Pl. 104, figs 25–30). In general, the valves of M. vietnamica sp. nov. have less convex valve margins. The axial area of M. fossalis is clear, narrow-lanceolate, more rarely linear, in the middle expanded roundish-lanceolate, differentiating a central area due to some shortened striae. The central area in M. vietnamica sp. nov., is well defined and transversely widened; it is surrounded by shortened striae. However, the striae in M. fossalis appear more strongly radiate than in M. vietnamica sp. nov. M. fossalis is the only species that has coarse areolae. Mayamaea fossalis and M. vietnamica sp. nov. have similar striae densities (16–21 in 10 μm in M. fossalis vs. 19–22 in 10 μm in M. vietnamica sp. nov.) and areolae (ca. 22 in 10 μm in M. fossalis vs. 20–30 in 10 μm in M. vietnamica sp. nov.) (Lange-Bertalot 2001). However, M. fossalis has a greater number of areolae in the stria (up to 3, 4, or 5) (Reichardt 2018, p. 170, Pl. 125, LM figs 42–51, Pl. 126, SEM figs 9 & 10), while in M. vietnamica sp. nov. there are no more than 2 or 3 areolae per stria. One SEM of the valve exterior of M. fossalis showed only 2 or 3 areolae per stria and striae slightly radiate, was published by Reichardt (2018, Pl. 126, fig. 8, with the specimen coming from a small stream in the war cemetery on Nagelberg Bächlein in der Kriegsgräberstätte am Nagelberg, Germany), and this specimen might be M. vietnamica sp. nov. Another SEM micrograph of an external valve, named as “Mayamaea cf. fossalis” by Barragán et al. (2017, SEM fig. 40, from Craste de Louley River at Lacanau, France), also probably corresponds to M. vietnamica sp. nov. Despite the morphological similarity, molecular genetic data do not show a close relationship between M. vietnamica sp. nov. and M. fossalis (Fig. 2).
Mayamaea recondita (Hustedt) Lange-Bertalot is morphologically similar to M. vietnamica sp. nov. in valve shape. There is no central area in M. recondita, while in M. vietnamica sp. nov. the central area is well defined. Moreover, the axial area of M. recondita is generally narrower than that of M. vietnamica sp. nov. Striae of M. recondita are biseriate, while in our species they are uniseriate, the density of areolae is highest among representatives of the genus (70–80 in 10 μm), in comparison with M. vietnamica sp. nov. (20–30 in 10 μm). Also, the number of areolae per striae is high (3–6 in M. recondita vs. 1–3 in M. vietnamica sp. nov.).
Mayamaea terrestris resembles M. vietnamica sp. nov. in the shape of axial area, which expands towards the valve center. However, M. terrestris differs by lacking a central area, whereas in M. vietnamica sp. nov. the central area is well defined. The density of striae of M. terrestris is generally higher (22–24 [26] in 10 μm vs. 19–22 in 10 μm in M. vietnamica sp. nov.), as well as the density of areolae (50 in 10 μm vs. 20–30 in 10 μm in M. vietnamica sp. nov.). The number of areolae in the striae in M. terrestris is also higher (1–6 against 1–3 in M. vietnamica sp. nov.).