Flora of drift plastics: a new red algal genus, Tsunamia transpacifica (Stylonematophyceae) from Japanese tsunami debris in the northeast Pacific Ocean
Article information
Abstract
Floating debris provides substrates for dispersal of organisms by ocean currents, including algae that thrive on plastics. The 2011 earthquake and tsunami in Tohuku, Japan resulted in large amounts of debris carried by the North Pacific Current to North America from 2012 to 2016. In 2015–2016, the plastics in the debris bore a complex biota including pink algal crusts. One sample (JAW4874) was isolated into culture and a three-gene phylogeny (psbA, rbcL, and SSU) indicated it was an unknown member of the red algal class Stylonematophyceae. It is a small pulvinate crust of radiating, branched, uniseriate filaments with cells containing a single centrally suspended nucleus and a single purple to pink, multi-lobed, parietal plastid lacking a pyrenoid. Cells can be released as spores that attach and germinate to form straight filaments by transverse apical cell divisions, and subsequent longitudinal and oblique intercalary divisions produce masses of lateral branches. This alga is named Tsunamia transpacifica gen. nov. et sp. nov. Sequencing of additional samples of red algal crusts on plastics revealed another undescribed Stylonematophycean species, suggesting that these algae may be frequent on drift oceanic plastics.
INTRODUCTION
The increase in plastics in the ocean is a major hazard for oceanic and coastal organisms (Derraik 2002, Barnes et al. 2009, Andrady 2011, Eriksen et al. 2014). Floating debris can also provide opportunities for long-distance dispersal of otherwise poorly dispersing organisms (Fraser et al. 2011, Cumming et al. 2014). Plastics harbor communities that appear to be specific to these substrates (Carson et al. 2013, Zettler et al. 2013). Diatoms and particular bacterial communities are often known to inhabit small fragments of the marine plastisphere (Bravo et al. 2011, Zettler et al. 2013, Samoray 2016).
The North Pacific Ocean is especially rich in plastic debris (Goldstein et al. 2013, Eriksen et al. 2014, Law and Thompson 2014). The highly destructive Tohoku earthquake and subsequent tsunami of Mar 11, 2011 deposited more than 5 million tons of debris into the ocean (Japanese Ministry of the Environment 2012). A part of this debris, including many plastic items, was buoyant enough to float and be carried by currents across the North Pacific to the coasts of Oregon and Washington, where they began arriving in June of 2012. Many debris items were recognizable as Japanese by their markings, manufacturing details, and fouling biota. The fouling biota of tsunami debris is now being investigated by a number of scientists (Carlton et al. 2016, Hansen et al. 2016), but studies of the specific organisms that survive on the plastic debris have just begun.
The red algal class Stylonematophyceae has about 15 genera (West et al. 2007, Zuccarello et al. 2008, Yoon et al. 2010, Guiry and Guiry 2016). Morphologically they are simple, varying from unicells and colonies to filamentous prostrate and upright thalli with various plastid types (single multilobed or multiple discoid plastids, with or without a pyrenoid). Most are marine genera but a few occur in freshwater and terrestrial habitats. Some appear to be ubiquitous on various other substrates (e.g., algae, plants, wood, nets, and glass) and do not have clear biogeographic patterns (Zuccarello et al. 2008). Seven genera (Bangiopsis, Chroodactylon, Chroothece, Colacodictyon, Rhodaphanes, Rhodosorus, and Stylonema) have a single multi-lobed plastid with a prominent pyrenoid whereas Empselium, Goniotrichopsis, Kyliniella, Neevea, Phragmonema, Purpureofilum, Rhodospora, and Rufusia have either a single complex multilobed plastid without a pyrenoid or several small discoid plastids without pyrenoids.
The reproductive modes clearly described for all Stylonematophyceae taxa are (1) cell division in unicells and colonies to produce spores (e.g., 4–32 spores per cell in Rhodospora) or (2) by release of vegetative cells directly as monospores in filamentous forms. Possible sexual reproduction was described once for the freshwater genus Kyliniella (Flint 1953); otherwise sexual structures have not been reported.
Within the Rhodophyta, two ultrastructural details are characteristic of the Stylonematophyceae: (1) plastids (except Rhodaphanes) have a peripheral encircling thylakoid (also seen in Florideophyceae and Compsopogonophyceae). In other classes, an encircling thylakoid is absent in plastids; (2) Golgi bodies are associated with the endoplasmic reticulum in the Stylonematophyceae and Compsopogonophyceae. In other classes of the Rhodophyta, Golgi bodies are associated with mitochondria (Scott et al. 2010).
As part of a broader study we are investigating the red algal crusts found on plastic debris in order to determine their identity and to compare them with other members of this diverse class of red algae.
MATERIALS AND METHODS
Specimens of crusts were collected from plastic debris items (Table 1) cast ashore on Oregon and Washington beaches during the winter and spring of 2015 and 2016, nearly 5 years after the Japanese tsunami. Several of the debris items contained Japanese writing that indicated their origin was the Tohoku coast of Japan. Nearly all of the debris supporting the alga was light in colour and determined to be high-density polyethylene (HDPE), a plastic that typically floats at the waterline. Specimens were preserved in 5% formalin in seawater for later photography and in silica gel for molecular analysis. One collection of the crust (GIH-416), found to be mostly unialgal, was kept alive in preliminary culture at 11°C using F/2 medium (Andersen 2005) until further investigations could be carried out. GIH-416 was taken from a white polyethylene bottle (Fig. 1A) found on Nov 5, 2015 on the Long Beach Peninsula in Washington (46°37.385 N, 124°04.246 W). This material was the main reference collection for our observations of the species, and subsamples of this collection have been used to prepare the type material.
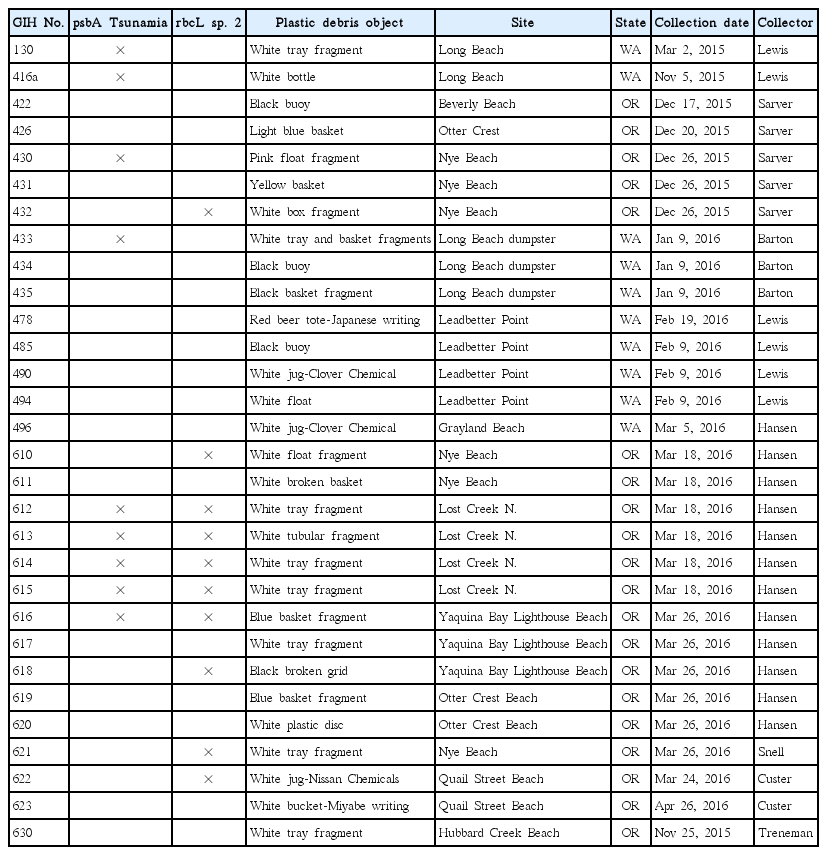
Samples of plastic debris collected on Oregon and Washington beaches in 2015 and 2016 containing Stylonematophyceae crusts

Tsunamia transpacifica (JAW4874). (A) Drift white polyethylene bottle from which the original specimens were scraped for culture. (B) Two-month-old crusts on glass cover slip showing typical radiating horizontally branched filaments with a dark pulvinate central mass of erect branches. Free spores and young sporelings visible around the crusts. (C) Most filaments show transverse divisions and one cell shows longitudinal division (double arrowhead). Enlarged cells (single arrowhead) with nucleus in the cell center. (D) Cell center with nucleus surrounded by particles and supported by thin radiating cytoplasmic strands. (E) Surface view of plastids with multiple lobes interconnected by wide and narrow strands. (F) Black starch grains (arrowheads) around nucleus of most cells treated with I2KI stain. (G) Central cell mass with cell divisions in different planes. Scale bars represent: A, 3 cm; B, 100 μm; C–G, 10 μm.
Living samples of GIH-416 were sent to Australia and a subculture (JAW4874) was placed in about 80 mL of low nutrient Modified Provasoli’s ES medium (5 mL per 1 L sterile seawater at 30‰ salinity) (West and McBride 1999) with 3 drops of 1% germanium dioxide to control diatoms and about 1 mg of powdered Na Penicillin G to control cyanobacteria. The 50 × 70 mm Pyrex dishes were maintained at 10 : 14 light-dark daily photoperiod, 19–22°C, and 4–8 μmol photons m−2 s−1 cool white LED lighting for several weeks while unialgal status, growth and reproduction were followed. A higher irradiance (12–34 μmol photons m−2 s−1) was used to observe morphological changes. To test for starch, filaments were placed on slides with drops of I2KI solution (Johansen 1940) and heated for 2 s at 800 W in a microwave oven.
All photographs except Fig. 1A were taken with a Zeiss GFL bright field microscope (Zeiss Australia, Sydney, Australia) and Canon G3 camera (https://www.canon.com.au/) and edited with Adobe Photoshop CS4 (http://www.adobe.com/au/) for publication.
Molecular analyses
Total DNA was isolated from silica gel-dried material using a modified CTAB procedure (Zuccarello and Lokhorst 2005). Amplification and sequencing of the plastid-encoded large subunit of the ribulose bisphosphate carboxylase/oxygenase gene (rbcL) used amplification primers, F8 (Wang et al. 2000) and F753-RrbcS-start (Freshwater and Rueness 1994). The psbA (photosystem II reaction center protein D1) gene was amplified using primers psbA-F and psbA-R1 (Yoon et al. 2002). Amplification of the nuclear SSU used a nested polymerase chain reaction (PCR): G01.1 (Müller et al. 2001) and SR12 (Nakayama et al. 1996) for the first round, and G01.1 and 18S-6M (Hanyuda et al. 2002), and LD7 (Kawai et al. 2005) and SR12 for the second round. Amplified products were checked for correct length, purity and yield in 1% agarose gels. PCR products were cleaned and commercially sequenced (Macrogen Inc., Seoul, Korea).
DNA sequences of members of the Stylonematophyceae and Compsopogonophyceae from previous publications were included (Zuccarello et al. 2008, 2011). Outgroups used in all analyses were Porphyridium aerugineum Geitler and Flintiella sanguinaria Ott, both taxa of the class Porphyridiophyceae. Sequences were edited, assembled and aligned using the Geneious software package, ver. 9.1 (Biomatters, available from http://www.geneious.com/). Independent trees were produced for each gene, and, because there was no hard incongruence between the data sets, a three-gene (rbcL, psbA, and SSU) concatenated dataset was produced for the culture isolate (JAW4874). Two other data sets for both the rbcL and the psbA genes were produced from PCR amplification and sequencing of DNA extracted from field samples. These amplification products gave sequences without apparent contamination (single peaks in electropherograms) but in some cases represented two organisms (see “Results”).
The program Modeltest ver. 3.7 (Posada and Crandall 1998) was used to find the model of sequence evolution that best fit the data set by an Akaike information criterion (AIC) (Posada and Crandall 2001). Maximum likelihood (ML) was performed with RAxML 7.2.8 (Stamatakis 2006). A saturation plot of the rbcL dataset, and not psbA, indicated that the third codon position for rbcL was saturated for this gene, therefore the third codon position was removed in the rbcL analysis. Data sets were partitioned by gene and codon, where appropriate. RAxML was performed with partitioned data and the GTR + gamma model and 1,000 non-parametric bootstrap replicates (Felsenstein 1985). Bayesian inference was performed with MrBayes v3.2 (Ronquist et al. 2012). Analyses consisted of two independent simultaneous runs of one cold and three incrementally heated chains, and 3 × 106 generations with sampling every 1,000 generations. The log files of the runs were checked with Tracer v1.5 (Rambaut and Drummond 2009) and a burn-in sample of 100 trees was removed from each run before calculating the majority rule consensus tree.
RESULTS
Field observations
Specimens of crusts were collected from plastic debris items (Table 1) cast ashore on Oregon and Washington beaches during the winter and spring of 2015 and 2016, nearly 5 y after the Japanese tsunami. Several of the debris items bore Japanese writing that indicated their origin was the Tohoku coast of Japan. Nearly all of the debris supporting the algae was light in colour and determined to be HDPE, a plastic that typically floats at the waterline. The items included small pieces of floats (12-cm in diameter), larger bottles (Fig. 1A), and large totes (160 cm or more in length). The red algal crusts were typically part of a small community of species that inhabited the plastic including diatoms, an Ulvella-like green alga, several hydroids, and Lepas anatifera Linnaeus 1758, the pelagic gooseneck barnacle.
Microscopic examination of field crusts revealed features characteristic of the red algal class Stylonematophyceae. The crusts were palmelloid with numerous single subglobose cells 8–10 μm in diameter, short filaments and small packets of cells enclosed in a clear mucilaginous sheath up to 500 μm thick. The chloroplasts lacked pyrenoids and appeared to be discoid or highly lobed. When debris items with crusts were stored in a dark cold room (5°C) for up to 6 months, the alga remained alive, retaining its colour and structural characteristics.
Culture observations
The live specimen in collection number 416 (culture number JAW4874) was placed in culture on glass cover slips. In two months, 0.3–1.0 mm pulvinate crusts were produced by radiating filaments with many lateral and vertical branches (Fig 1B). Each vegetative cell had one spherical central nucleus about 3 μm in diameter, suspended on thin cytoplasmic strands radiating outward across the central vacuole to the peripheral cytoplasm. Small granules around the nucleus and in the strands (Fig. 1C, D & G) stained black with I2KI, a typical starch reaction (Fig. 1F, arrowheads). Some cells were not stained. The single parietal plastid had many variously shaped lobes interconnected by thin to wide strands (Fig. 1E). No pyrenoid was visible.
Crusts scraped from the substrate and transferred to fresh medium released numerous spores within 2 h (Fig. 2A). Vegetative cells in dense pulvinate masses formed abundant spores via numerous oblique and longitudinal divisions (Fig. 1B & G). No pit connections were visible between any cells. Spores were 10 (9–12) μm diam, with a central nucleus and a single, lobed parietal plastid. Free spores showed no motility and usually adhered to the substrate less than 12 h after release. Attached spores formed 2-celled (about 10 × 20 μm) and 4-celled (about 10 × 30–40 μm) stages by apical transverse divisions (Fig. 2B). In the horizontal young filaments 8–9 cells long (about 10–12 × 65–75 μm), mid-section intercalary cells initiated longitudinal and oblique divisions to form lateral branches (Fig. 2B). The apical cells of filaments continued to divide transversely and filaments radiated horizontally outward. In the middle section of the main axial filament and in lateral branches, longitudinal and oblique cell divisions produced erect branches (Fig. 2C) resulting in compact three-dimensional crusts up to 1 mm wide and many cells high.
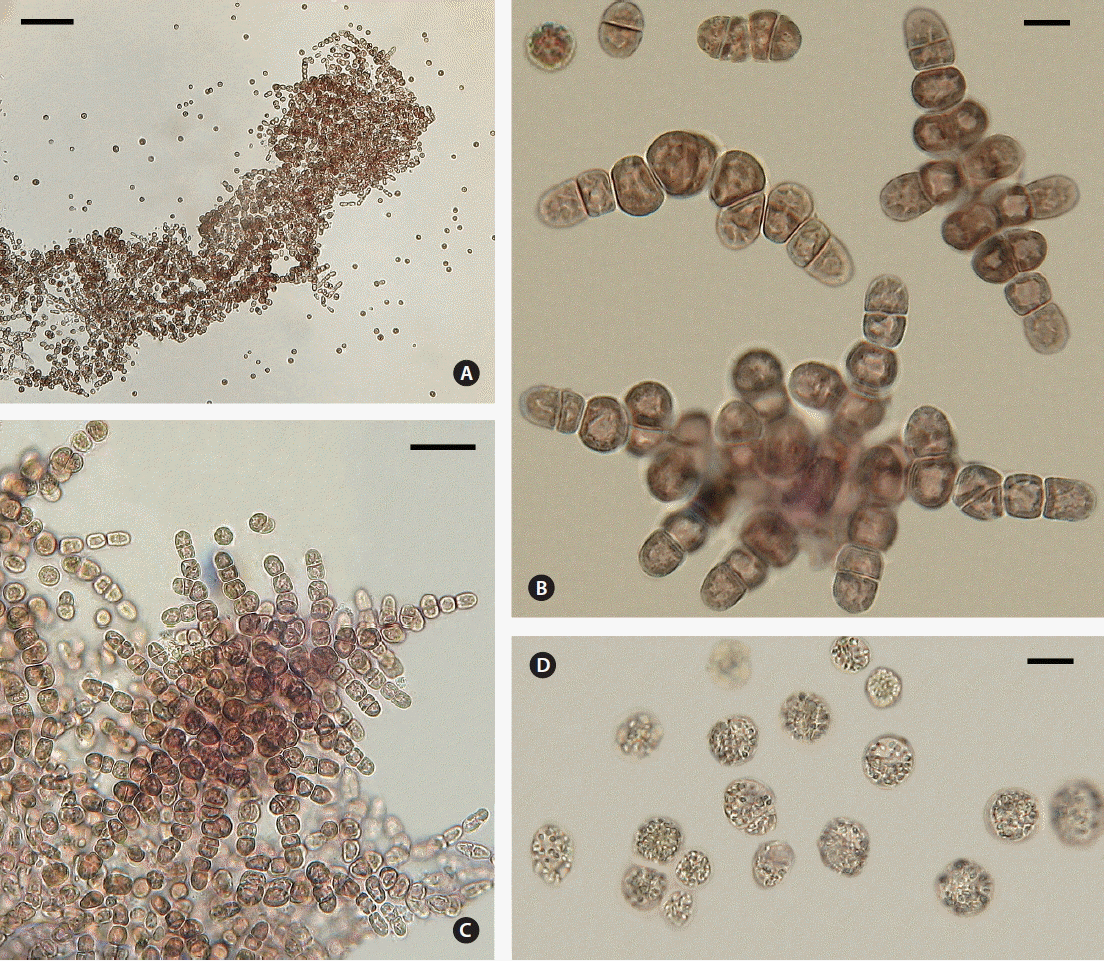
Tsunamia transpacifica (JAW4874). (A) Many spores released two hours after crust removed from glass cover slip. (B) Series of cell divisions of main filament on glass: 2 cells; 4 cells; 8-celled main filament with 4 middle cells showing oblique and transverse divisions; 9-celled main filament with 4 intercalary cells forming lateral and upright branching by transverse and oblique divisions; 15-celled main filament with 10 intercalary cells forming lateral and upright branching by transverse and oblique divisions, as terminal cells of filaments continue to divide transversely. (C) Well-developed crust with extensive horizontal and vertical branching. (D) Film at air/water interface, single and double cells, pale with many cytoplasmic granules, probably starch. Scale bars represent: A, 100 μm; B & D, 10 μm; C, 80 μm.
In stationary cultures with bright light (12–30 μmol photons m−2 s−1), spores sometimes rose to the air/water interface, forming floating films up to 2 cm wide and one or more cells thick. In these films spores divided, producing separate single cells and 2-celled filaments (Fig. 2D). The cells were pale, with reduced photosynthetic pigments, and contained many large cytoplasmic granules, probably starch, a typical response to nutrient deficiency in red algae.
Molecular analysis
A three-gene data set (SSU, rbcL, and psbA) indicated that the algal isolate JAW4874 is different from any known species in the Stylonematophyceae (Fig. 3). The three-gene phylogeny was not supported in many early branches and did not support grouping this isolate with any previous member of the class. Because the new sequences did not match known genera, the morphological characters did not match known species and the habitat of this alga was unusual, we propose a new genus and species for the entity isolated as JAW4874.
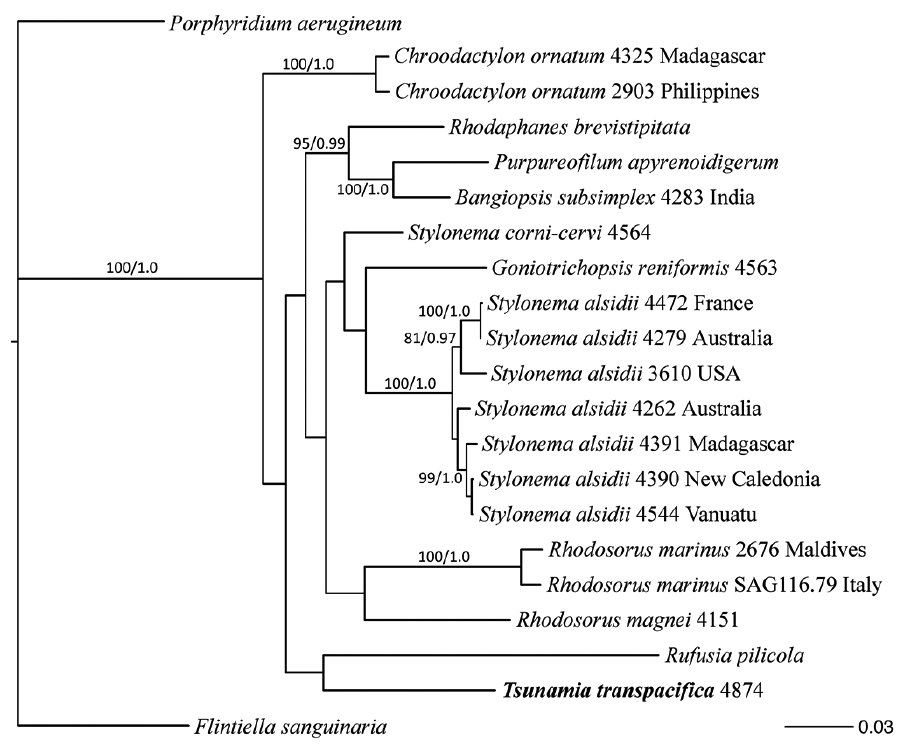
Maximum-likelihood (ML) phylogenetic topology of a three-gene data set (rbcL, psbA, and SSU) of various Stylonematophyceae. Support values at each node are bootstrap (BP) values in percent from RAxML (ML BP) (left) and Bayesian posterior probability (PP) (right). Values <70% ML BP and <0.95 PP removed. Outgroups used were Porphyridium aerugineum and Flintiella sanguinaria.
Amplification using psbA primers from field samples showed that Tsunamia transpacifica was found in eight different samples of plastics from Washington and Oregon, USA (Table 1). These sequences from field specimens were identical to each other and identical to sequences from the cultured T. transpacifica (Fig. 4, dataset of 885 bp) and did not form a clade with any other species in the Stylonematophyceae.
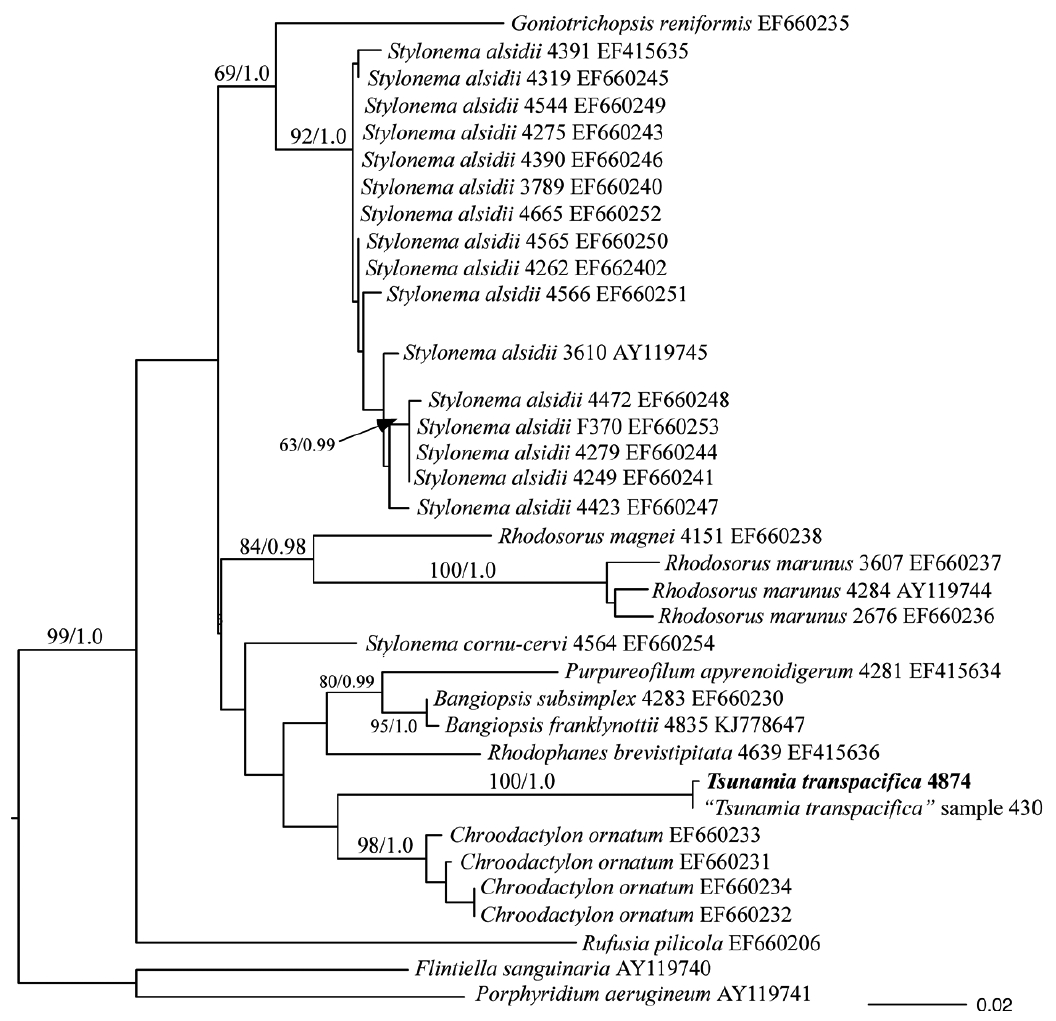
Maximum-likelihood (ML) phylogenetic topology of psbA sequence data of various Stylonematophyceae. All samples of Tsunamia transpacifica were identical. Support values at each node are bootstrap (BP) values in percent from RAxML(ML BP) (left) and Bayesian posterior probability (PP) (right). Values <70% ML BP and <0.95 PP removed. Outgroups used were Porphyridium aerugineum and Flintiella sanguinaria.
Amplification of rbcL from field samples (10 samples, dataset of first and second positions, 810 bp) amplified a different species of Stylonematophyceae (Fig. 5). While the position of these sequences within the Stylonematophyceae was not well supported, it is clear that they are different from T. transpacifica and are not similar to any other sequences available. This new entity was fairly common on plastics. In some DNA extracts (5 field samples), both entities were recovered, one using psbA primers (T. transpacifica) and the other using rbcL primers (Stylonematophyceae). Because we did not notice any morphologically distinct specimens from field-collected samples, we suspect that this species is probably morphologically similar to T. transpacifica, but until it is placed in unialgal culture, this cannot be determined. The alga described in the field observations above may be either a new species or something different, in view of the various molecular results.

Maximum-likelihood (ML) phylogenetic topology of rbcL sequence data of various Stylonematophyceae and Compsopogonophyceae. New undescribed Stylonematophyceans in bold. Support values at each node are bootstrap (BP) values (ML BP) in percent from RAxML (left) and Bayesian posterior probability (PP) (right). Values <70% ML BP and <0.95 PP removed. Outgroups used were Porphyridium aerugineum and Flintiella sanguinaria.
Tsunamia gen. nov. J. A. West, G. I. Hansen, G. C. Zuccarello et T. Hanyuda
Description
In culture, vegetative cells are directly released as single non-motile spores (9–12 μm diam.) attaching to a glass substrate and germinating to form radiating, branched uniseriate filaments that become dense pulvinate crusts up to 1mm wide that are often confluent. Horizontally radiating filaments are formed by apical cells undergoing transverse divisions, followed by longitudinal and oblique divisions of intercalary cells to form numerous horizontal and erect branched filaments. No pit connections form between cells. Each cell has one spherical central nucleus about 3 μm diameter, suspended on thin cytoplasmic strands radiating outward across the central vacuole and a single, peripheral, purple to pink, much-lobed plastid without a pyrenoid. Identity was also based on the basis of SSU, rbcL, and psbA sequences.
Tsunamia transpacifica sp. nov. J. A. West, G. I. Hansen, G. C. Zuccarello et T. Hanyuda
Description
Species description as for genus.
Holotype
The holotype culture specimen was deposited in the Royal Botanic Gardens Melbourne, National Herbarium of Victoria, Australia (MEL 2399992). Strain JAW4874 (field collection number 416) was from a white plastic bottle found in debris of the Mar 11, 2011 Tohoku earthquake and tsunami, obtained on Nov 5, 2015 on the Long Beach Peninsula, Washington, USA (46°37.385 N, 124°04.246 W) by Russ Lewis (Table 1). Isotype culture specimens were also deposited in the University of Michigan Herbarium (MICH 701751) and University Herbarium, University of California, Berkeley (UC 2050560). It was not possible to designate a single alga in the field collection as the type. Consequently, a single thallus was reisolated from the laboratory culture in Melbourne and selected as the holotype and isotypes.
Culture deposit
The culture (JAW4874) has been accessioned (CCMP 3460) by the Provasoli-Guillard National Center for Marine Algae and Microbiota (NCMA), Bigelow Laboratory for Ocean Sciences, P.O. Box 380, 60 Bigelow Drive, East Boothbay, ME 04544, USA.
Etymology
The name reflects the origin of the type specimen, obtained from a plastic bottle transported in tsunami drift across the Pacific Ocean to the west coast of North America.
Genbank accession numbers
SSU, KX787935; psbA, KX787937; rbcL, KX787936.
DISCUSSION
The discovery and description of this species was facilitated by several events and discoveries of recent years. The Japanese 2011 tsunami produced large quantities of marine debris that floated across the North Pacific to wash ashore in Oregon and Washington where material could easily be collected. A portion of the debris consisted of HDPE, a plastic widely used in various products. Since this plastic floats at the waterline, it provided a good substrate and habitat for light-requiring species in the Stylonematophyceae. Recent studies on the life history and molecular biology of the Stylonematophyceae (West et al. 2005, 2007, 2014) gave us the necessary background and framework for our recognition of the species as new.
The simple morphology with few diagnostic characters of the Stylonematophyceae makes specimen identification difficult without molecular evidence. Definitive identification also requires isolation into unialgal culture combined with cytological and chemical observations. Tsunamia is superficially similar to the endozoic Stylonematophycean Neevea repens Batters which has horizontal palmelloid or pseudo-filamentous thalli and several small discoid plastids per cell (Garbary et al. 1980b) unlike the single multilobed plastid of Tsunamia. Neevea has never been cultured nor is molecular data available for this genus.
Without molecular evidence, Tsunamia might have been identified as a member of the Erythropeltidales (Compsopogonophyceae) (e.g., Pseudoerythrocladia), because of its horizontal pulvinate thallus with multi-branched radiating filaments, although spores in the Erythropeltidales form by oblique divisions of vegetative cells (Garbary et al. 1980a, Zuccarello et al. 2010, West et al. 2012). rather than by oblique and longitudinal divisions, as in Tsunamia. Furthermore, in contrast to spores of other Stylonematophyceae observed in culture (Pickett-Heaps et al. 2001, Wilson et al. 2002, West et al. 2005, 2007, 2012, 2014), neither gliding nor amoeboid motility was observed in Tsunamia spores.
Our initial surveys of plastic debris indicate that members of the red algal class Stylonematophyceae thrive on floating plastics (Table 1). With the increasingly widespread use of both polyethylene and polypropylene in a wide array of items that often end up in debris, the new “plastisphere” environment in the world’s oceans (Zettler et al. 2013, Samoray 2016) continues to increase in size, providing ample habitat for species that thrive on plastic. The Stylonematophyceae asexual life cycle and high spore production aid in the maintenance of their populations, and their floating plastic substrates disperse the populations widely via the prevailing currents. The pink crusts on plastic debris included two different members of the Stylonematophyceae. One of these, Tsunamia transpacifica, was isolated in culture and characterised as a new genus, while the other is only known as a DNA sequence. It is likely that additional cryptic molecular species will be found in this habitat with future studies.
Where did the populations of these two species on plastic debris originate? Did they exist in Japan on marine structures or in near-shore areas before the tsunami, colonizing debris, and drifting across the Pacific to North America? Did they attach to the plastic in the open ocean? Or did they attach to the debris in the near-shore regions of North America? It is difficult to say because the species have not been observed previously in either Japan or North America. On plastic debris, they co-exist with populations of species that are known to be pelagic, including hydroids and Lepas anatifera. It is possible that the species either attached to the debris via pelagic spores in the water column (Amsler and Searles 1980) or that they hitch-hiked on pelagic hydroids known to drift in the water column (Concelman et al. 2001). Once on the plastic debris, they could survive for long periods of time, propagating repeatedly and jumping between objects that are in close proximity, such as those that might occur in the North Pacific garbage patch. Where did Tsunamia occur before the development of plastic? It may have been a pelagic species that became an opportunistic primary colonizer of the new “plastisphere” in the North Pacific.
It is intriguing that both T. transpacifica and the unidentified Stylonematophycean were found with different sets of primers. The only explanation is variation in the primer binding sites for these two species, with higher specificity in T. transpacifica for the psbA primer combination, while the unidentified Stylonematophyceans seem to be more readily amplified with the rbcL primer pairs. Continued study and tests of alternative primers will help to determine the distribution and relative abundance of these two species on plastic debris.
This investigation highlights our lack of knowledge of the algal diversity found on this ubiquitous floating ocean debris, with potential effects on native algal floras. Continued study of floating algae, possibly on plastics from the central Pacific gyre or from attached plastics in Japan, may determine if these algae have a Japanese origin or if they are native to pelagic substrates.
ACKNOWLEDGEMENTS
We thank Russ Lewis and John Chapman who made the initial collections of the debris objects. The Japanese Ministry of the Environment through the North Pacific Marine Science Organization (PICES) provided partial support for GIH & TH during the current study. The US Environmental Protection Agency provided laboratory space for GIH during the study. JW used personal funds for culturing and light microscopy and is grateful to Geoff McFadden, University of Melbourne, for use of facilities and supplies in doing this research. Susan Loiseaux de Goër provided critical Photoshop advice for arranging and improving the photographic figures. For accessioning Tsunamia herbarium specimens, we thank Nimal Karunajeewa and Pina Milne, the Royal Botanic Gardens, Melbourne; Michael Wynne, University of Michigan; and Kathy Ann Miller, University of California at Berkeley. Julie Sexton accessioned the culture at CCMP, Bigelow Laboratory for Ocean Sciences, Maine.