Morphological observations and phylogenetic position of the parasitoid nanoflagellate Pseudopirsonia sp. (Cercozoa) infecting the marine diatom Coscinodiscus wailesii (Bacillariophyta)
Article information
Abstract
During a sampling at Nokdong harbor, southern coast of Korea in January 2017, the marine diatom Coscinodiscus wailesii cells infected by a novel parasitoid nanoflagellate were observed. While the development process of the trophosomes of the parasitoid was more similar to that of Pseudopirsonia mucosa, division pattern of the auxosomes was similar to that of Pirsonia species. Phylogenetic analyses inferred from 18S rRNA gene sequences revealed that the parasitoid infecting C. wailesii fell within the cercozoan groups and branched as a sister lineage of the clade consisting of Pseudopirsonia mucosa and the undescribed Cercomonas sp. SIC7235, with the sequence dissimilarity of 7.3% with Pseudopirsonia mucosa. All of these developmental and molecular characteristics suggest that the parasitoid nanoflagellate infecting the diatom C. wailesii is a new Pseudopirsonia species.
INTRODUCTION
Planktonic diatoms are one of key primary producers as well as one of the dominant phytoplankton in aquatic ecosystems. They are almost constantly confronted with parasites in aquatic environment and are susceptible to infections by a variety of eukaryotic parasitoids (i.e., which always kill their hosts to complete their life cycles), including cercozoans, chytrids, dinoflagellates, euglenoids, oomycetes, and stramenopiles (e.g., Drebes 1966, Drebes and Schnepf 1988, 1998, Kühn et al. 1996, Tillmann et al. 1999, Bulman et al. 2001). Among those parasitoids, in particular, species belonging to the genus Pirsonia have been well documented as parasitoids infecting a number of marine diatoms (Schnepf et al. 1990, Kühn et al. 1996, Schweikert and Schnepf 1997). The Pirsonia species share some morphological characteristics of the life cycles, including formation of trophosome and auxosome. Typical infections of the parasitoids initiate when the motile flagellate attaches to the host diatom frustule and then penetrates into the host cell using a pseudopodium. The pseudopodium inside the host cell becomes a trophosome that digests host protoplasm in a food vacuole and transports the digested host materials to the auxosome, a remaining part of the parasitoid flagellate on the outside host cell. Seven species in the genus, Pirsonia diadema, P. formosa, P. eucampiae, P. guinardiae, P. mucosa, P. punctigerae, and P. verrucosa, have been so far described only from the North Sea (Schnepf et al. 1990, Kühn et al. 1996, 2004). Recent molecular phylogenetic analyses demonstrated that P. mucosa was very distantly related from other Pirsonia species. While other Pirsonia species clustered within stramenopiles forming a monophyletic clade, P. mucosa diverged within the heterogenic group of cercomonad (Kühn et al. 2004). For this reason along with additional morphological characteristics, P. mucosa was recently moved to a new genus Pseudopirsonia by Kühn et al. (2004).
The parasitoid nanoflagellates Pirsonia/Pseudopirsonia species are known to display a different degree of host specificity and host range from laboratory cross infection experiments and field observations (Kühn et al. 1996, 2004). For example, P. diadema, P. punctigerae, P. guinardiae, and P. verrucosa are relatively host-specific. While P. diadema and P. punctigerae infect only diatoms of the genera Coscinodiscus and Thalassiosira, respectively, the latter two parasitoids (P. guinardiae and P. verrucosa) infect only species belonging to the genus Guinardia. By contrast, P. formosa and Pseudopirsonia mucosa have a broad host range and they could successfully parasitize various diatom species across several host genera. Nonetheless, the nonspecific P. formosa and Pseudopirsonia mucosa did not infect the tested Coscinodiscus species (i.e., C. concinnus, C. granii, and C. wailesii) (Kühn et al. 1996, 2004). So far, the sole Pirsonia species known to be capable of infecting Coscinodiscus species, including C. wailesii, is P. diadema.
During sampling at Nokdong harbor of Korea in January 2017, the marine diatom Coscinodiscus wailesii cells infected by a novel parasitoid nanoflagellate were encountered. Here, we presented the developmental morphological characteristics of the parasitoid, relative to those of previously described P. diadema. In addition, the molecular phylogenetic analyses based on18S rRNA gene sequences were performed to determine the phylogenetic affiliation of the novel parasitoid nanoflagellate to other Pirsonia/Pseudopirsonia species.
MATERIALS AND METHODS
Sampling and cell isolation
Concentrated seawater samples were collected using a 20 μm plankton net from Nokdong harbor, Korea (34°31′26.74″ N, 127°8′8.38″ E) on Jan 11, 2017 and transported to the lab for further examination. Water temperature and salinity were measured using a Yellow Spring Instrument (YSI, Yellow Springs, OH, USA). The diatom C. wailesii cells infected by the parasitoid nanoflagellates were individually isolated using a capillary pipet under an inverted microscope (Axio Vert 1, Carl Zeiss Inc., Göttingen, Germany), washed in serial drops of filtered seawater, and transferred to six-well plates containing 4 mL of filtered seawater. The well plates were placed on shelf of an incubator at 20°C on a 14 : 10 light : dark cycle, with cool-white fluorescent lamps providing 100 μmol photons m−2 s−1 and examined at every 12-h interval.
Light microscopy
Specimens were observed using an inverted microscope (Axio Vert A1, Carl Zeiss Inc., Oberkochen, Germany) with differential interference contrast optics. Light micrographs were taken at 100×−400× magnification using a Full HD mini box camera (MediCAM-X, Comart System, Seoul, Korea) photomicrographic system coupled with the microscope (Kim et al. 2015).
DNA extraction, polymerase chain reaction, and sequencing
About 150 cells of parasitoid nanoflagellates detached from the host cells at the late stage of infection were collected using a capillary pipet, washed several times with sterile filtered seawater, placed into polymerase chain reaction (PCR) tubes, and finally pelleted by centrifugation. Total genomic DNA was extracted from the pellets using a chelex extraction method (Kim and Park 2014). The 18S rRNA gene region of the parasitoid nanoflagellate was amplified using primer sets: Euk328f and Euk329r (Moon-Van Der Staay et al. 2000). PCRs were performed in a total of 20 μL of reaction solution containing 3 μL of DNA (chelex extract) as a template using an AccuPower PCR premix kit (Bioneer, Daejeon, Korea). The reactions were conducted using a C1000 Touch Thermal Cycler (Bio-Rad, Hercules, CA, USA) with the following conditions: initial denaturing step at 95°C for 4 min followed by 40 cycles (95°C for 20 s, 55°C for 20 s, and 72°C for 1 min), with a final extension at 72°C for 5 min. Amplified products were visualized on EcoDye (SolGent Co., Daejeon, Korea) stained 1% agarose gels, purified by a PCR purification kit (Bioneer, Daejeon, Korea) and sequenced with primers (Euk328f, Euk329r, Euk516r, and Euk1209r) using a Big-Dye Terminator v3.0 Cycle Sequencing kit (Applied Biosystems, Foster City, CA, USA) and an ABI model 3730 sequencer (Applied Biosystems), according to manufacturer’s protocols. The amplicons were sequenced until at least double stranded coverage was reached. ContigExpress (Vector NTI ver. 10.1; Invitrogen, Grand Island, NY, USA) was used to edit out low quality regions and assemble the sequence reads. The assembled sequences were verified by comparison using BLASTN search in the NCBI database and deposited in GenBank (accession number MF615236).
Alignments and phylogenetic analyses
Sequences were primarily aligned using CLUSTALX 1.83 (Larkin et al. 2007) and were further refined manually using MacClade 4.08 (Maddison and Maddison 2000). Unambiguously aligned positions were selected and were applied for phylogenetic analyses. Modeltest v.3.7 (Posada and Crandall 1998) was used to select the most appropriate model of substitution for the maximum likelihood (ML) method. GTR + I + Γ (i.e., general time reversible with invariant sites and gamma rate correction) model was identified as the best-fit model for 18S rDNA dataset. ML analyses were performed using RAxML 8.0.0 with the general time-reversible model with gamma correction and 1,000 replicates (Stamatakis 2014). Bayesian analysis used MrBayes 3.1.1 (Ronquist et al. 2012) running four simultaneous Monte Carlo Markov Chains for 2,000,000 generations and sampling every 100 generations, following a prior burn-in of 100,000 generations (1,000 sampled trees were discarded). A consensus tree was constructed from 19,001 post burn-in trees.
RESULTS AND DISCUSSION
Morphological features
During the sampling of marine diatom Coscinodiscus wailesii parasitized by a novel parasitoid nanoflagellate, water temperature and salinity were 8°C and 32, respectively. Although the diatom Rhizosolenia setigera was predominant and various other diatoms also co-occurred in the samples, infections by the parasitoid nanoflagellates were observed only on C. wailesii. The infected C. wailesii cells were easily distinguishable, due to the appearance of “a diadem” (Figs 1 & 2). Such an apparent appearance was similar to that of the parasitoid nanoflagellate Pirsonia diadema infecting the marine diatoms Coscinodiscus spp. from the North Sea near Helgoland. The Coscinodiscus cells heavily infected by P. diadema also displayed the appearance of a “diadema” in that every rimoportulae forming a ring at the margin of the diatom valve was occupied by attachment of the parasitoids (Kühn et al. 1996).
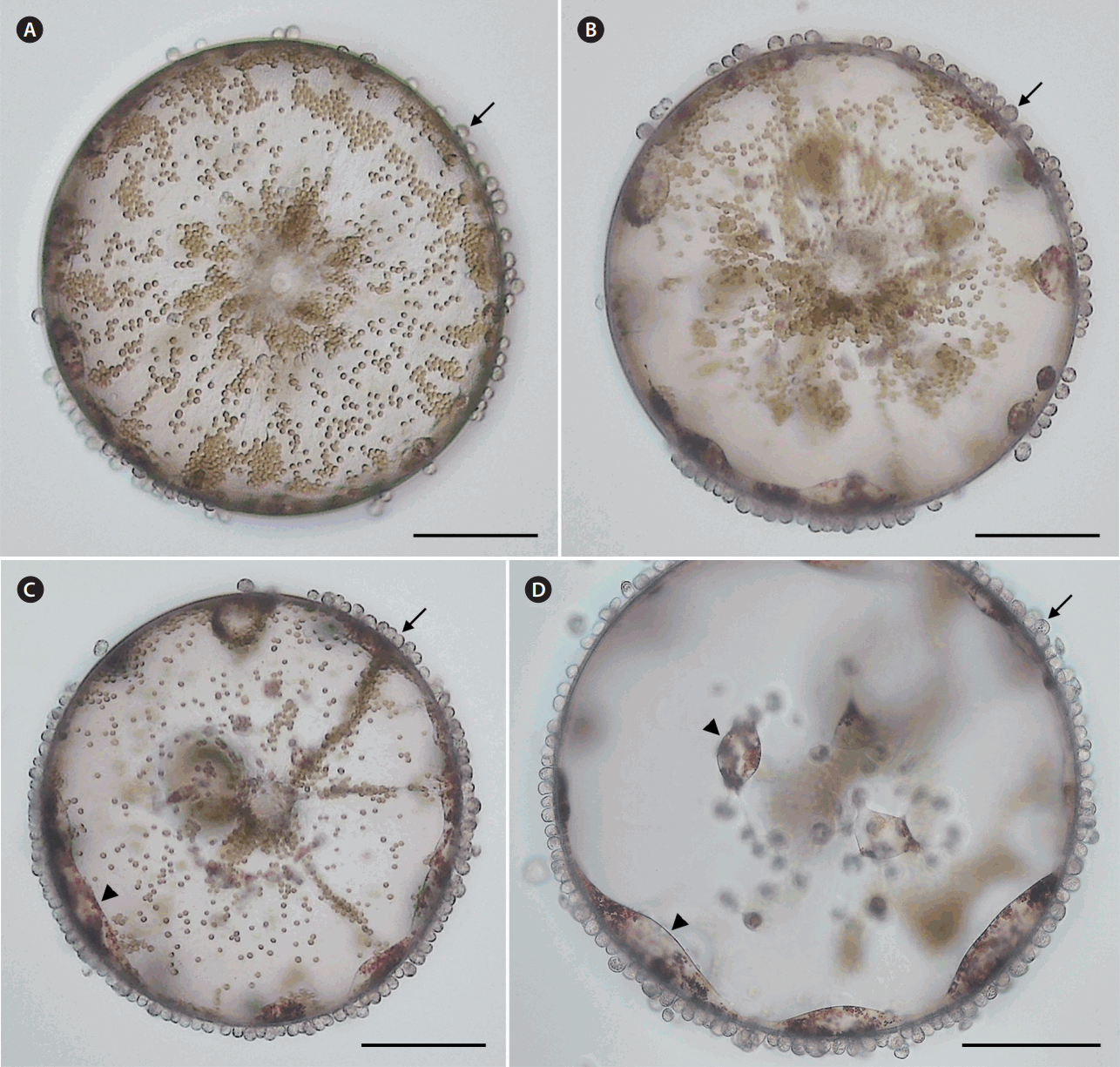
The new parasitoid nanoflagellate Pseudopirsonia sp. infecting the diatom Coscinodiscus wailesii. Time series light microscopic images of the same infected C. wailesii cell just after isolating from the field sample (A), at 12-h (B), 24-h (C), and 36-h (D) incubations. Note the numerous auxosomes (arrows) increasing over time at the margin of the diatom valve and lateral large trophosomes (arrowheads) formed by fusion with adjacent trophosomes inside the host diatom. Scale bars represent: A–D, 100 μm.
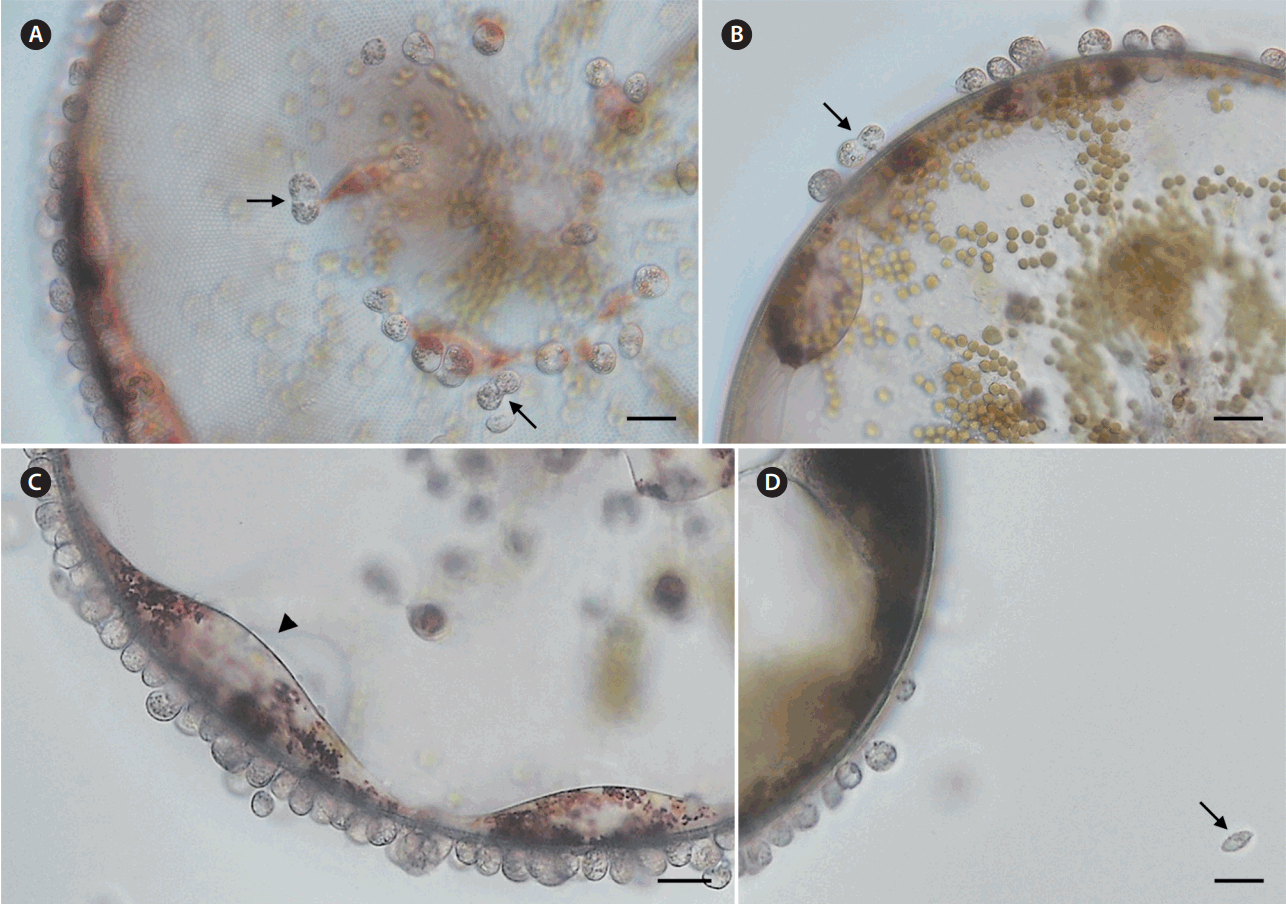
Light micrographs of Pseudopirsonia sp. infecting the diatom Coscinodiscus wailesii. (A & B) Auxosomes at the margin of the host diatom valves and valve faces. Arrows indicate the first division of the primary auxosomes of the parasitoid. (C) Auxosomes and trophosomes of the parasitoids. Arrowhead indicates fused trophosomes of the parasitoids. (D) The motile stage of the parasitoid. Arrow indicates the mature flagellate. Scale bars represent: A–D, 20 μm.
Microscopic observations of live infected C. wailesii cells individually isolated from the field samples at every 12-h interval (Fig. 1A–D) revealed that the number of parasitoid nanoflagellates gradually increased over time, with the host protoplast being ingested and almost completely consumed after 36 h (Fig. 1D). Infections by the novel parasitoids were mostly observed at the margin of the diatom valve, but also additionally found on the valve face (Fig. 2A). Once the motile flagellate attached to the host, its flagella disappeared during the feeding stage (Fig. 2B). The attached flagellate penetrated into the frustule of the host diatom using a pseudopodium, which later became a trophosome inside the diatom, with some part of the flagellate, which became an auxosome, still remained outside the host cell. The trophosomes gradually ingested the host protoplast phagocytotically and fused with several adjacent trophosomes as growing over time (Fig. 2C). This developmental process of trophosomes did more resemble with that of Pseudopirsonia mucosa rather than other Pirsonia species (Kühn et al. 2004). Pseudopirsonia mucosa attaches to the diatom frustule and forms an unusually broad pseudopod that is situated laterally, while other Pirsonia species attach with a posteriorly protruded pseudopod (Kühn et al. 2004).
The auxosomes of the novel parasitoid had a globular shape with 12 ± 0.2 μm (mean ± SE, n = 40) in diameter (Fig. 2A–C). The size of the auxosomes in the new parasitoid was similar to that of other Pirsonia species ranging from 10 to 15 μm, but smaller than that of Pseudopirsonia mucosa with 18 μm in diameter (Kühn et al. 2004). The auxosomes in the new parasitoid divided longitudinally and the resulting daughter cells appeared to remain connected with trophosomes (Fig. 2A & B). Such division pattern of the auxosomes of our Pseudopirsonia sp. was more similar to that of other Pirsonia species rather than Pseudopirsonia mucosa, in which its auxosomes divided as a morula-shape and covered by a mucilaginous coat (Kühn et al. 1996, 2004).
Mature flagellates of the new parasitoid Pseudopirsonia sp. had an elliptical shape and were flattened laterally with the size of 7.3 ± 0.2 μm × 14.4 ± 0.6 μm (mean ± SE, n = 3) (Fig. 2D). Their movement showed a slowly gliding pattern (personal observation). These morphological feature and movement pattern of the motile flagellate does more resemble to Pseudopirsonia mucosa because the motile flagellates of Pseudopirsonia has an oval-oblong shape and slowly gliding movement, unlike Pirsonia spp. having a rounded to oval shape and slightly jerking swimming movement (Kühn et al. 1996, 2004).
Phylogenetic analyses
Partial 18S rRNA gene sequences of the novel parasitoid collected from two infected C. wailesii cells were obtained and all sequences (1,726 nucleotides in length) of the isolates were identical. BLAST search of Genbank provided a 92% maximum match of the novel parasitoid sequence to those of several cercozoan genera including Pseudopirsonia mucosa (AJ561116), Protaspis spp. (FJ824122–FJ8224125), Cryothecomonas longipes (AF29040), Thaumatomastix sp. (GQ144681), and Allas sp. (AY268040).
Phylogenetic analyses inferred from 18S rRNA gene sequences revealed that the parasitoid Pseudopirsonia sp. infecting C. wailesii fell within the cercozoan groups and branched as a sister lineage of the clade of Pseudopirsonia mucosa and the undescribed Cercomonas sp. SIC7235 with high statistical supports of bootstrap proportion (BP)/posterior probabilities (PP) (91/1.0) (Fig. 3). The marine sand-dwelling cercozoan Clautriavia biflagellata showed a sister relationship of Pseudopirsonia sp. with moderate statistical supports of BP/PP (76/1.0). Pairwise comparison of the partial 18S rDNA sequences showed 122 base differences between Pseudopirsonia sp. and Pseudopirsonia mucosa based on 1,669 unambiguously aligned sites with 7.3% dissimilarity. By comparison, all species in the genus Pirsonia formed a monophyly with robust statistical supports of BP/PP (100/1.0) and placed within stramenopiles in the 18S rRNA gene tree (Fig. 3). The Pirsonia species were very closely related to each other with showing low dissimilarity of only 0.2–2.4% (Kühn et al. 2004). The best trees generated with ML and Bayesian methods were largely congruent and in those trees the Pirsonia species diverged into two distinct lineages, one composing of three P. formosa strains and P. diadema and the other including P. punctigerae and the clade of P. verrucosa and P. guinardiae, although inner nodes for the relationships were weakly to moderately supported (Fig. 3).

RAxML phylogenetic tree inferred from 1,885 unambiguously aligned sites of 18S rDNA sequences including 47 stramenopiles and 43 cercozoan of ingroup taxa and two sequences of glaucophytes as outgroup taxa. Numbers shown on nodes are support values of bootstrap percentages using RAxML fast bootstrapping analysis and Bayesian posterior probabilities higher than 60% and 0.6, respectively. Black circles indicate robust statistical supports of bootstrap proportion/posterior probabilities (100/1.0). Open circles represent 1.0 of posterior probabilities.
CONCLUSION
The parasitoid nanoflagellate Pseudopirsonia sp. infecting the marine diatom Coscinodiscus wailesii presented in this study was unique compared to other Pirsonia/Pseudopirsonia species previously described in some ways. While development process of the trophosome was more similar to that of Pseudopirsonia mucosa, division pattern of the auxosome was similar to that of Pirsonia species. Furthermore, phylogenetic analyses based on 18S rRNA gene sequence revealed that the parasitoid Pseudopirsonia sp. fell within cercozoa group instead of stramenopiles containing other Pirsonia species. The new parasitoid nanoflagellate was closely related to Pseudopirsonia mucosa, but showed 7.3% sequence dissimilarity. All of these developmental and molecular characteristics suggest that the parasitoid nanoflagellate infecting the diatom C. wailesii is a new Pseudopirsonia species. Unfortunately, however, failure to establish the host diatom-parasite in culture precluded further close examinations of the parasitoid, including detailed morphological features of the motile flagellate and its host range. Further studies are needed to identify the potential new parasitoid observed in this study and better understand its autecology, as well as to investigate the diversity of the species within the genus Pseudopirsonia, in which only one species has been reported.
ACKNOWLEDGEMENTS
This work was supported by a Research Grant of Pukyong National University (C-D-2016-1156) and research grants funded by the National Research Foundation of Korea (NRF-2014R1A2A1A11053911 and 2017R1A2B4002658) to SK and (NRF-2016R1A6A1A03012647) to MGP.