Five Alexandrium species lacking mixotrophic ability
Article information
Abstract
Mixotrophy in marine organisms is an important aspect of ecology and evolution. The discovery of mixotrophic abilities in phototrophic dinoflagellates alters our understanding of the dynamics of red tides. In the phototrophic dinoflagellate genus Alexandrium, some species are mixotrophic, but others are exclusively autotrophic. There are differences in the ecological roles of autotrophic and mixotrophic Alexandrium in marine food webs. However, of the 34 known Alexandrium species, the mixotrophic ability of >20 species has yet to be explored. In this study, the mixotrophic capabilities of Alexandrium insuetum CCMP2082, Alexandrium mediterraneum CCMP3433, Alexandrium pacificum CCMP3434, Alexandrium tamutum ATSH1609, and Alexandrium margalefii CAWD10 were investigated by providing each species with 22 diverse prey items including bacterium-sized microbeads (1 μm), the cyanobacterium Synechococcus sp., algal prey species, and the ciliate Mesodinium rubrum. None of the 5 Alexandrium species fed on any of the prey items. These results increase the number of Alexandrium species lacking mixotrophic abilities to 9, compared to the 7 known mixotrophic Alexandrium species. Furthermore, the Alexandrium phylogenetic tree based on the large subunit ribosomal DNA contained 3 large clades, each of which had species with and without mixotrophic abilities. Thus, the acquisition or loss of mixotrophic abilities in Alexandrium might readily occur.
INTRODUCTION
Mixotrophy is defined as a trophic mode combining photosynthesis and phagotrophy within a single organism and is present in a diversity of protistan groups, including dinoflagellates and ciliates (Jeong et al. 2010, Hansen 2011, Johnson 2011, Stoecker et al. 2017). Mixotrophic protists are ubiquitous, inhabiting all areas from tropical to polar seas (Mitra et al. 2016, Stoecker et al. 2017, Faure et al. 2019, Leles et al. 2019). Mixotrophy is important not only because of its significance to the evolution of protists but also because of its ecological role in marine ecosystems. Mixotrophy is thought to have been a crucial event in the evolution of endosymbiosis because the plastids in most eukaryotes originate from the phagotrophic ingestion and retention of unicellular photosynthetic organisms (Raven 1997, Jones 2000). Moreover, the presence or absence of mixotrophy in phototrophic species alters our understanding of its role in marine food webs (Stoecker 1999, Burkholder et al. 2008, Jeong et al. 2012, 2015), the cycling of oceanic elements (Ward and Follows 2016), and biogeochemical models (Mitra et al. 2016). Mixotrophic species play multiple roles as primary producers, prey, and predators, whereas autotrophic species are exclusively primary producers and / or prey (Jeong et al. 2010). Thus, exploring the mixotrophic ability of phototrophic species is a critical step in understanding the function of marine ecosystems.
Dinoflagellates are a major component of marine ecosystems (Falkowski et al. 2004, Simon et al. 2009, Jeong et al. 2013). They often form red tides or harmful algal blooms (HABs) that may cause human illness and the large-scale mortality of fin-fish and shellfish (Anderson et al. 2012, Park et al. 2013, Jeong et al. 2017, Glibert et al. 2018). Thus, their ecological and physiological characteristics (i.e., trophic mode, growth, and feeding) receive much attention. Over the past two decades, research has shown that many phototrophic dinoflagellates that were thought to be exclusively autotrophic are mixotrophic (Bockstahler and Coats 1993, Jacobson and Anderson 1996, Stoecker et al. 1997, Berge et al. 2008, Jeong et al. 2016, Lim et al. 2018). It has been suggested that most HAB causative species are mixotrophic (Flynn et al. 2018), but less than 5% of the phototrophic dinoflagellates (approximately 40 species among ca. 1,200 phototrophic species) have been categorized as mixotrophic (Jeong et al. 1997, 2016, Hansen 2011, Blossom et al. 2012, Gómez 2012, Lee et al. 2014, 2016, Lim et al. 2015, 2018). Therefore, many more phototrophic dinoflagellates need to be tested for mixotrophic abilities.
Among phototrophic dinoflagellates, those in the genus Alexandrium are major HAB species (Hallegraeff 1995, Landsberg 2002, Anderson et al. 2012, Davidson et al. 2016). Many Alexandrium species can cause large-scale fish mortality and / or human illness by producing toxins, so scientists and members of the aquaculture industry have paid close attention to their dynamics (Cembella et al. 2002, Tillmann et al. 2008, Anderson et al. 2012, Grattan et al. 2016). To minimize the harmful effects of Alexandrium, it is important to understand the ecophysiology of each species and predict its population dynamics. Moreover, the establishment of models to predict HAB dynamics has revealed that the trophic mode of the HAB species is very important–some species enhance their growth rates by mixotrophy (Jeong et al. 2015). Furthermore, among the 34 known Alexandrium species (Guiry and Guiry 2019), only 13 have been examined for mixotrophy (Jeong et al. 2005a, 2005b, Yoo et al. 2009, Blossom et al. 2012, 2017, Lim et al. 2015, Lee et al. 2016). To better predict and model Alexandrium blooms, it is necessary to explore the trophic modes of more species.
In the present study, the mixotrophic abilities of 5 Alexandrium species, Alexandrium insuetum CCMP2082, Alexandrium mediterraneum CCMP3433, Alexandrium pacificum CCMP3434, Alexandrium tamutum ATSH1609, and Alexandrium margalefii CAWD10 were investigated. The mixotrophic abilities of A. mediterraneum, A. pacificum, and A. tamutum have never been explored, whereas the mixotrophic abilities of A. insuetum and A. margalefii have been investigated, but only with one prey species, Teleaulax acuta (Blossom et al. 2017). Thus, A. insuetum and A. margalefii need to be tested with additional prey items. This study used 22 diverse prey items, including bacterium-sized microbeads, the cyanobacterium Synechococcus sp., algal prey species, and the ciliate Mesodinium rubrum. All of these prey items are from taxa commonly found in marine environments and have been used in prior mixotrophy tests of phototrophic dinoflagellates (Jeong et al. 2004, 2016, Lee et al. 2014, 2016, Lim et al. 2015, 2018, Jang et al. 2017). The results presented here add to our understanding of the roles of Alexandrium species in marine ecosystems and dinoflagellate evolution.
MATERIALS AND METHODS
Preparation of experimental organisms
Diverse prey items were provided as potential prey (Table 1). The bacterium-sized microbeads (1 μm in diameter; Fluoresbrite Microspheres, Polysciences, Inc., Warrington, PA, USA) were diluted into freshly filtered seawater and sonicated for 1 min before use to avoid agglomerated particles. The cyanobacterium Synechococcus sp. and all algal prey species, except Margalefidinium polykrikoides and Lingulodinium polyedra, were grown at 20°C in enriched f/2 (Guillard and Ryther 1962) or L1 (National Center for Marine Algae and Microbiota, East Boothbay, ME, USA) seawater media under cool white fluorescent lights (20 μmol photons m−2 s−1) on a 14: 10 h light / dark cycle. Cells of M. polykrikoides and L. polyedra were grown under continuous illumination (50 μmol photons m−2 s−1) with cool white fluorescent lights because they do not grow well under lower illumination on a light / dark cycle (Lee et al. 2014). The mixotrophic ciliate M. rubrum was maintained by offering Teleaulax amphioxeia as prey every week. The mean equivalent spherical diameters (ESDs) of the phytoplankton species were obtained from previous studies (Jeong et al. 2015, 2016, Lee et al. 2016).
A clonal culture of A. tamutum ATSH1609 was established from Shiwha Bay, Korea (Kang et al. 2018). The clonal cultures of A. insuetum CCMP2082, A. mediterraneum CCMP3433, and A. pacificum CCMP3434 were obtained from the National Center for Marine Algae and Microbiota, USA. The clonal culture of A. margalefii CAWD10 was obtained from the Cawthron Institute’s Culture Collection of Microalgae, New Zealand. As the concentrations increased, the cultures were sequentially transferred to 50, 270, and 800 mL flasks containing fresh f/2-Si or L1 seawater media. All of the flasks were placed on a shelf at 20°C under cool white fluorescent lights (20 μmol photons m−2 s−1) on a 14: 10 h light / dark cycle.
Exploring the mixotrophic abilities of Alexandrium species
The experiment was designed to investigate if the Alexandrium species can feed on each target potential prey species when a diet of diverse prey items was provided (Table 1).
A dense culture of each Alexandrium species was transferred to a 500 mL polycarbonate (PC) bottle containing f/2-Si or L1 medium. Triplicate 1 mL aliquots were then removed and examined under a compound microscope to determine the concentrations of each Alexandrium species.
Approximately 4.2 × 107 bacterium-sized fluorescent beads were added into one 42-mL PC bottle, each containing a target Alexandrium species. One ‘bead only’ control bottle and one Alexandrium control bottle (without added beads) were set up for each experiment. The bottles were filled with freshly filtered seawater, capped, and placed on a vertically rotating plate (0.9 r min−1), and incubated at 20°C under a 14: 10 h light / dark cycle at 20 μmol photons m−2 s−1 illumination. At the beginning, and after 2, 24, and 48 h incubation periods, 5-mL aliquots were removed from each bottle, transferred into a confocal dish, and then examined under an epifluorescence microscope (Zeiss-Axiovert 200M; Carl Zeiss Ltd., Göttingen, Germany) at ×100–630 magnification. After subsampling, the bottles were again filled with freshly filtered seawater, capped, and placed on a vertically rotating plate, and incubated.
When Synechococcus sp. was used as prey, the initial concentrations of Alexandrium species and Synechococcus sp. (ca. 2 × 106 cells mL−1) were established using an auto pipette to deliver a predetermined volume of culture with a known cell abundance to the experimental bottles. One 80-mL PC experimental bottle and one predator control bottle were set up with a single prey concentration for each Alexandrium species. The bottles were incubated and a 5-mL aliquot was removed from each bottle and fixed in formalin (final concentration = 3%) at the intervals as described above. The fixed aliquots were filtered onto 5-μm pore-sized, 25 mm PC black membrane filters, and the concentrated cells on the membranes were then observed under an epifluorescence microscope with green light excitation at ×1,000 magnification to determine whether the predator was able to feed on Synechococcus sp.
The initial concentrations of the tested Alexandrium species and each target prey species were established using an auto pipette to deliver a predetermined volume of culture with a known cell density to the experimental bottles. One 42 mL PC bottle, with mixtures of the Alexandrium species and target potential prey and predator control bottles containing the tested Alexandrium only, were set up for each target prey species. The bottles were filled with freshly filtered seawater, capped, and placed on a vertically rotating plate (0.9 r min−1), and incubated under the conditions described above. After 2, 24, and 48 h, a 5 mL aliquot was removed from each bottle and transferred into 6-well chamber plates. After subsampling, the bottles were again filled with freshly filtered seawater, capped, and placed on a vertically rotating plate, and incubated. To determine if the Alexandrium species were able to feed on the target potential prey species, >20 cells were tracked to examine the physical contact, attack (attempt to capture), and successful capture (ingestion) with a dissecting microscope (SZX2-ILLB; Olympus, Tokyo, Japan) at ×10–63 magnification. Furthermore, the lysis and immobilization of the target potential prey cells was also investigated. Cells of the tested Alexandrium species and each target prey species were photographed on slides with cover glasses using a digital camera (Zeiss AxioCam HRc 5; Carl Zeiss Ltd.) on the light and epifluorescence microscopes at ×200–630 magnification. Moreover, Teleaulax amphioxeia cells after being incubated with the tested Alexandrium species for 1 h were also photographed using the digital camera on the inverted microscope at ×400 magnification.
DNA sequencing and phylogenetic analysis
The genomic DNA (gDNA) sequence of A. tamutum ATSH1609 has never been reported, whereas the sequences for A. insuetum CCMP2082, A. mediterraneum CCMP3433, A. pacificum CCMP3434, and A. margalefii CAWD10 have been published. Thus, the gDNA sequence of A. tamutum ATSH1609 was analyzed in the present study. Furthermore, to establish phylogenetic trees, the gDNA sequences of Alexandrium affine AATA1308, Alexandrium andersonii AAJH1505, and Alexandrium fraterculus AFYS1309, which were previously investigated for mixotrophic abilities (Lee et al. 2016), were also analyzed here.
To obtain the gDNA sequences of each Alexandrium species, a 10 mL aliquot of each dense culture was transferred to a 15 mL tube and concentrated at 6,000 ×g for 1 min. The pellet was then used for the gDNA extraction. The gDNA was extracted using an AccuPrep Genomic DNA Extraction Kit (Bioneer, Daejeon, Korea). The amplification reaction mixtures (50 μL in total volume) were as follows: 5 μL of 10× F-StarTaq buffer, 1 μL of 10 mM of dNTP mix, 0.25 μL of 5 U/μL BioFACT F-Star Taq DNA polymerase (BioFACT Co., Ltd., Daejeon, Korea), 2 μL of 10 μM of each primer, and 1 μL of the extracted gDNA. The primers used to amplify regions of large subunit ribosomal DNA (LSU rDNA) are D1R (5′-ACCCGCTGAATTTAAGCATA-3′) (Scholin et al. 1994), 28–1483R (5′-GCTACTACCACCAAGATCTGC-3′) (Daugbjerg et al. 2000), and LSUB (5′-ACGAACGATTTGCACGTCAG-3′) (Litaker et al. 2003). The gDNA was amplified in a Mastercycler EP Gradient thermal cycler (Eppendorf, Hamburg, Germany) using the following cycling conditions: 3 min at 95°C followed by 40 × 45 s at 95°C, 1 min at the selected annealing temperature (AT), and 1 min at 72°C, with a final extension of 5 min at 72°C. The AT was adjusted depending on the primers used, according to the manufacturer’s instructions. Positive and negative controls were used for all amplification reactions. The PCR products were purified using an AccuPrep DNA Purification Kit (Bioneer), and sequencing was performed with an ABI 3730XL DNA Analyzer (Applied Biosystems, Foster City, CA, USA). The obtained sequences were deposited on GenBank.
DNA sequence alignment for the phylogenetic analyses of the LSU rDNA region of the Alexandrium species were conducted using the software MEGA 4 (Tamura et al. 2007), including the sequences of closely related taxa obtained from GenBank. Maximum likelihood (ML) analysis of the region was conducted using the program RAxML 7.0.3 with the default GTRGAMMA model (Stamatakis 2006). Further, 200 independent tree inferences were used to identify the best tree. ML bootstrap values were determined using 1,000 replicates. Bayesian analysis was conducted using MrBayes v.3.1 (Huelsenbeck and Ronquist 2001, Ronquist and Huelsenbeck 2003) with the default GTR + G + I model to determine the best available model for the data. For the sequence region, four independent Markov Chain Monte Carlo runs were performed, as described in Kang et al. (2010).
RESULTS
Mixotrophic abilities of Alexandrium spp
Alexandrium insuetum CCMP2082, A. mediterraneum CCMP3433, A. pacificum CCMP3434, A. tamutum ATSH1609, and A. margalefii CAWD10 did not feed on any of the bacterium-sized microbeads, cyanobacterium Synechococcus sp., algal prey species, or M. rubrum (Table 1, Figs 1 & 2). Furthermore, none of the five Alexandrium species showed any attacking behavior toward the prey species.
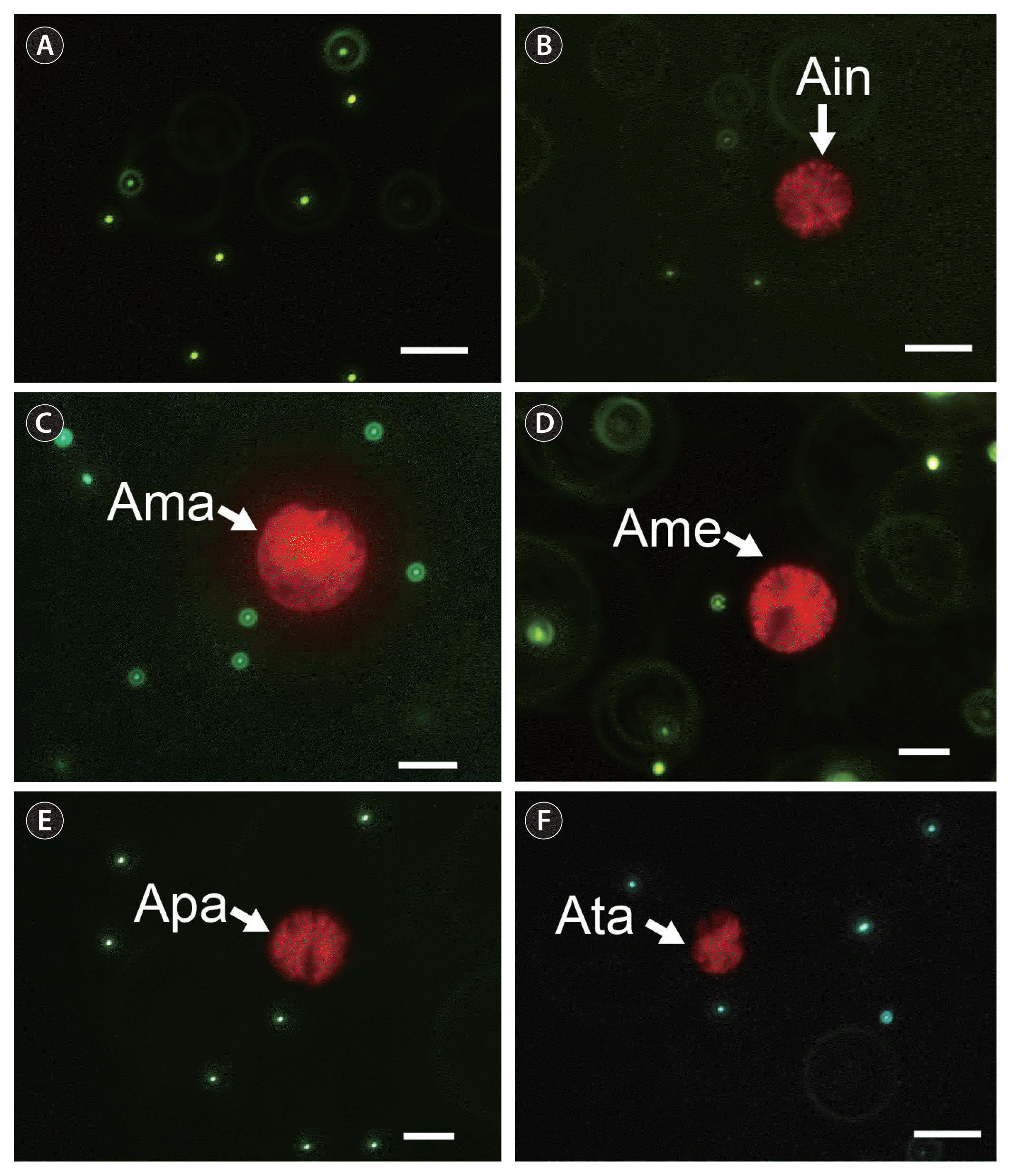
Micrographs of the bacterium-sized microbeads (1 μm, green color) (A) and the target Alexandrium species (white arrow) incubated with the microbeads for one day (B–F), taken under an epifluorescence microscope. (B) Alexandrium insuetum CCMP2082 (Ain). (C) A. margalefii CAWD10 (Ama). (D) A. mediterraneum CCMP3433 (Ame). (E) A. pacificum CCMP3434 (Apa). (F) A. tamutum ATSH1609 (Ata). No bead was found inside the protoplasm of the target Alexandrium species cell. Scale bars represent: A–F, 20 μm.
Micrographs of the target Alexandrium species (green and white arrows) incubated with Amphidinium carterae (Ac, blue and yellow arrowheads) that was provided as prey, taken under light (A, C, E, G, I) and epifluorescence microscope (B, D, F, H, J). (A & B) Alexandrium insuetum CCMP2082 (Ain). (C & D) Alexandrium margalefii CAWD10 (Ama). (E & F) Alexandrium mediterraneum CCMP3433 (Ame). (G & H) Alexandrium pacificum CCMP3434 (Apa). (I & J) Alexandrium tamutum ATSH1609 (Ata). Scale bars represent: A–J, 10 μm.
A. insuetum, A. margalefii, A. mediterraneum, and A. pacificum lysed several algal and ciliate prey species, but A. tamutum did not lyse any of the prey (Table 1, Fig. 3). The number and kind of the algal and ciliate prey species that A. insuetum, A. margalefii, A. mediterraneum, and A. pacificum lysed differed from one another (Table 1); A. margalefii lysed 11 prey species, A. pacificum lysed 2 prey species (the cryptophytes T. amphioxeia and Storeatula major), and A. insuetum and A. mediterraneum each lysed 6 prey species.

Teleaulax amphioxeia (Ta) cells after being incubated without (A) and with (B–F) Alexandrium tamutum ATSH1609, Alexandrium insuetum CCMP2082, Alexandrium margalefii CAWD10, Alexandrium mediterraneum CCMP3433, and Alexandrium pacificum CCMP3434 for 1 h. (A) Intact Ta cells (blue arrowheads). (B) Ta cells (blue arrowheads) were not lysed by A. tamutum (Ata, green arrow). (C) Ta cells (red arrowheads) were lysed by A. insuetum (Ain, green arrow). (D) A Ta cell (red arrowhead) was lysed by A. margalefii (Ama, green arrow). (E) Several Ta cells (red arrowheads) were lysed by A. mediterraneum (Ame, green arrow). (F) Two Ta cells (red arrowheads) were lysed by A. pacificum (Apa, green arrows). Scale bars represent: A–F, 10 μm.
T. amphioxeia and S. major were lysed by all of the Alexandrium predator species except for A. tamutum, but Pyramimonas sp. and M. rubrum were lysed by A. insuetum, A. mediterraneum, and A. margalefii (Table 1). The relatively large dinoflagellates M. polykrikoides, Akashiwo sanguinea, and L. polyedra were lysed by only A. margalefii, whereas the relatively small dinoflagellate Prorocentrum donghaiense was lysed by only A. mediterraneum. The cryptophyte Rhodomonas salina and the relatively small dinoflagellate Amphidinium carterae were not lysed but were immobilized by A. insuetum (Table 1).
Phylogenetic analysis
In the phylogenetic tree based on the LSU rDNA region, A. insuetum CCMP2082, A. mediterraneum CCMP3433, A. pacificum CCMP3434, A. tamutum ATSH1609, and A. margalefii CAWD10 was positioned in clades representing each species (Fig. 4). A. pacificum and A. mediterraneum formed a large clade with A. affine, Alexandrium catenella, Alexandrium hiranoi, Alexandrium pseudogonyaulax, and Alexandrium tamarense, all of which were previously tested for mixotrophic abilities (Fig. 4). Furthermore, A. margalefii also formed a clade with Alexandrium pohangense (previously tested for mixotrophic ability). Moreover, A. insuetum and A. tamutum formed a big clade with Alexandrium minutum, Alexandrium ostenfeldii, and A. andersonii, all of which had also been previously tested for mixotrophy. In each of these 3 clades, Alexandrium species with and without mixotrophic abilities were present.
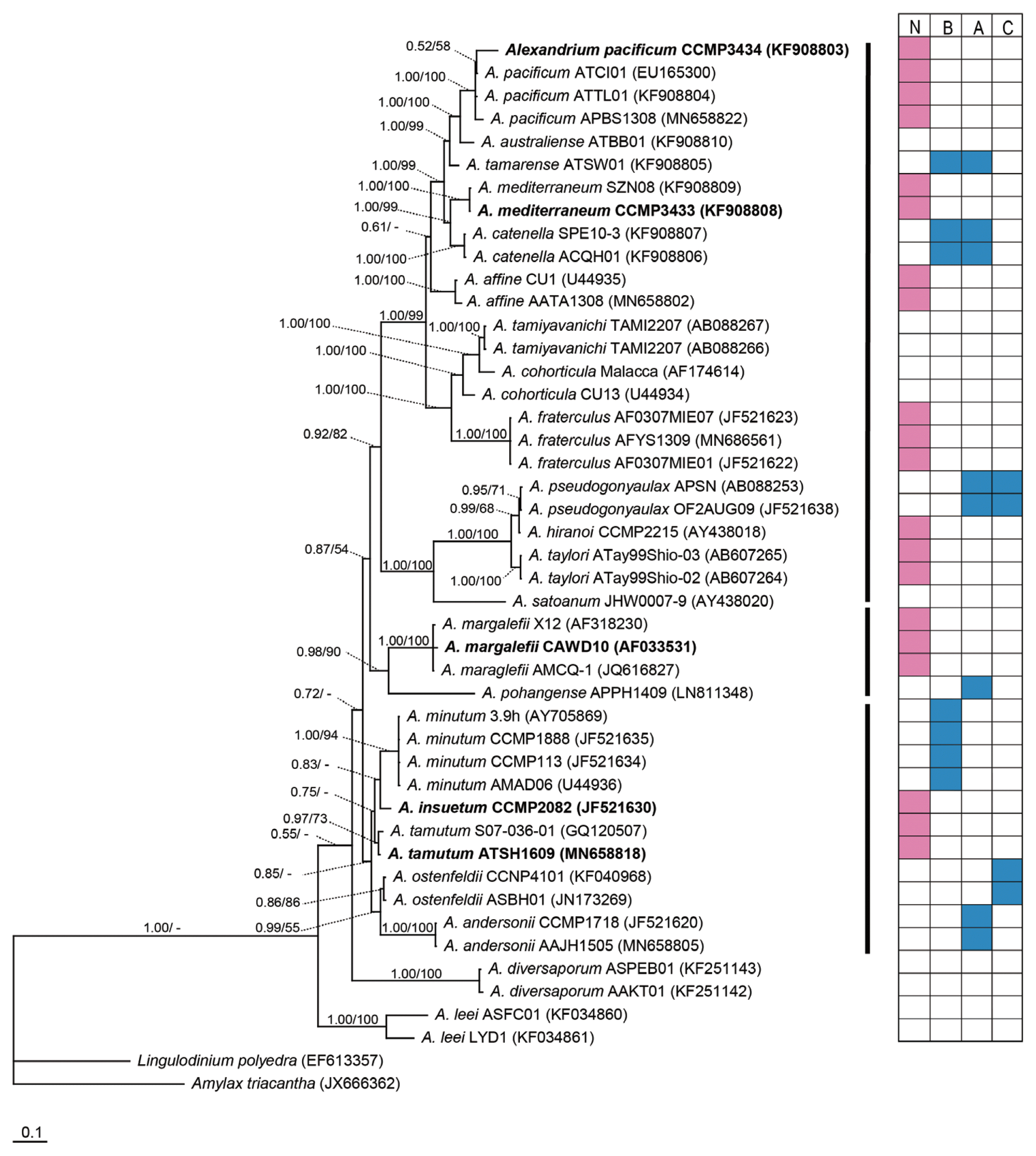
Consensus Bayesian tree based on 819 bp aligned positions of the large subunit ribosomal DNA region, using the GTR + G + I model and Amylax triacantha and Lingulodinium polyedra as outgroups. The parameters were as follows: assumed nucleotide frequencies as equal, substitution rate matrix with A–C substitutions = 0.1168, A–G substitutions = 0.2174, A–T substitutions = 0.1195, C–G substitutions = 0.0753, C–T substitutions = 0.3534, G–T substitutions = 0.1177, proportion of sites assumed to be invariable = 0.0807, and the rates for variable sites assumed to follow a gamma distribution with shape parameter = 0.8331. The branch lengths are proportional to the amount of character changes. The numbers above the branches indicate the Bayesian posterior probability (left) and maximum likelihood bootstrap values (right). Posterior probabilities ≥ 0.5 are shown. The tested strains in the present study were indicated in bold. N, no or lack of a mixotrophic ability; B, able to feed on bacteria; A, able to feed on algal prey; C, able to feed on ciliates. Data on the presence or lack of a mixotrophic ability of the Alexandrium species in the box were obtained from this study, Blossom et al. (2012, 2017), Jacobson and Anderson (1996), Jeong et al. (2005a, 2005b, 2015), Lee et al. (2016), Lim et al. (2015), and Yoo et al. (2009).
DISCUSSION
The results of this study clearly show that Alexandrium insuetum CCMP2082, Alexandrium mediterraneum CCMP3433, Alexandrium pacificum CCMP3434, Alexandrium tamutum ATSH1609, and Alexandrium margalefii CAWD10 lack mixotrophic abilities. Of the 13 previously tested Alexandrium species, the number with mixotrophic abilities (7) was slightly greater than those lacking mixotrophic abilities (6) (Fig. 5); A. andersonii, A. minutum, A. ostenfeldii, A. pohangense, A. tamarense, A. catenella, and A. pseudogonyaulax have mixotrophic abilities, whereas A. affine, A. fraterculus, Alexandrium taylorii, A. insuetum, A. margalefii, and A. hiranoi lack mixotrophy (Jacobson and Anderson 1996, Jeong et al. 2005a, 2005b, Yoo et al. 2009, Blossom et al. 2012, 2017, Lim et al. 2015, Lee et al. 2016). The results presented here confirm the lack of mixotrophic abilities in A. insuetum and A. margalefii, and brings the total number of Alexandrium species lacking mixotrophic abilities to 9. However, the lack of mixotrophic abilities in A. taylorii, and A. hiranoi should be confirmed with more prey species because only 1–4 prey species were tested in the previous study (Blossom et al. 2017). Still, the mixotrophic abilities of 18 Alexandrium species have yet to be explored. The presence or absence of mixotrophy in Alexandrium species will affect its ecological roles in different marine food webs; those lacking mixotrophic abilities act as primary producers and prey, while those species with mixotrophic abilities can also function as predators. In models predicting the outbreak, persistence, and decline of red tides or HABs by Alexandrium species, the growth rate (k) and mortality rate due to predation (g) are two critical parameters (Jeong et al. 2015, Lim et al. 2017). The growth rate of Alexandrium species lacking mixotrophic abilities is mainly affected by light intensity and nutrient concentrations, while the growth rate of mixotrophic Alexandrium species is also influenced by prey availability. Furthermore, mixotrophic Alexandrium species add to the mortality rate of prey species, causing red tides or HABs. Alexandrium species lacking mixotrophic abilities do not add to the mortality rate. Thus, it is necessary to determine the mixotrophic ability of target Alexandrium species to better inform the models of red tides or HABs.
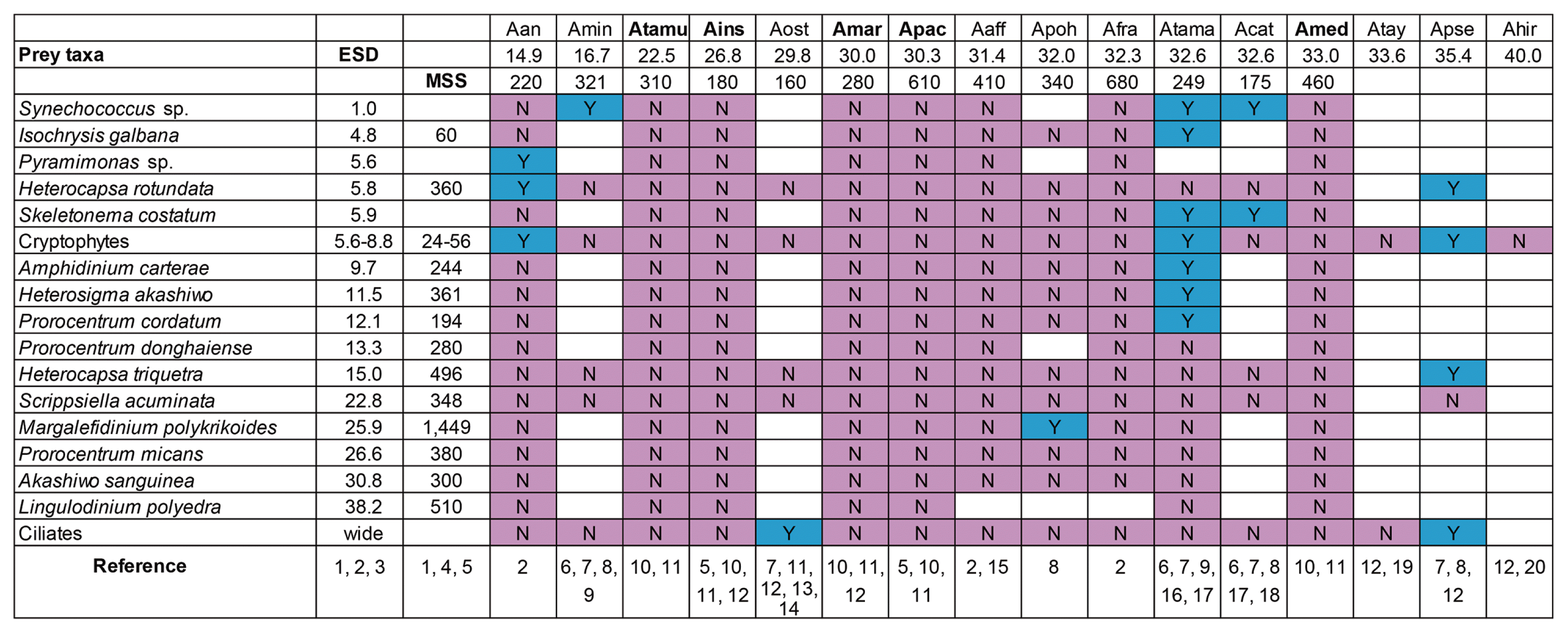
Feeding occurrence by each Alexandrium species on diverse prey items. Y in the blue box, the target Alexandrium predator species was observed feeding on the target prey cell; N in the pink box, the Alexandrium species was not observed feeding on the target prey cell. Blank box indicates a combination that was not tested. ESD, mean equivalent spherical diameter (μm); MSS, maximum swimming speed (μm s−1), Alexandriuim species in bold were tested in this study; Aan, Alexandrium andersonii; Amin, Alexandrium minutum; Atamu, Alexandrium tamutum; Ains, Alexandrium insuetum; Aost, Alexandrium ostenfeldii; Amar, Alexandrium margalefii; Apac, Alexandrium pacificum; Aaff, Alexandrium affine; Apoh, Alexandrium pohangense; Afra, Alexandrium fraterculus; Amed, Alexandrium mediterraneum; Atama, Alexandrium tamarense; Acat, Alexandrium catenalla; Atay, Alexandrium taylorii; Apse, Alexandrium pseudogonyaulax; Ahir, Alexandrium hiranoi. Reference: 1, Jeong et al. (2015); 2, Lee et al. (2016); 3, Jeong et al. (2016); 4, Skovgaard and Hansen (2003); 5, Lim et al. (2018); 6, Jeong et al. (2005a); 7, Blossom et al. (2012); 8, Lim et al. (2015); 9, Lewis et al. (2006); 10, this study; 11, Kang et al. (2018); 12, Blossom et al. (2017); 13, Jacobson and Anderson (1996); 14, Cembella et al. (2000); 15, Fraga et al. (1989); 16, Jeong et al. (2005b); 17, Yoo et al. (2009); 18, Karp-Boss et al. (2000); 19, Garcés et al. (1998); 20, Kita and Fukuyo (1988).
In the phylogenetic tree based on the LSU rDNA region, each of the 3 clades contained both Alexandrium species having and lacking mixotrophic abilities (Fig. 4). Thus, the acquisition or loss of a mixotrophic ability in Alexandrium might readily occur. Future studies should explore the direction of these changes, i.e., whether autotrophic species acquired mixotrophic abilities or whether mixotrophic species lost mixotrophic abilities and became autotrophic.
Among the 16 Alexandrium species tested for mixotrophic abilities, the two smallest species A. andersonii and A. minutum (14.9–16.7 μm in ESD) are mixotrophic, whereas the larger species A. insuetum and A. tamutum (22.5–26.8 μm) lack mixotrophic abilities (Fig. 5). A. tamarense, A. catenella, and A. pseudogonyaulax (32.6–35.4 μm) have mixotrophic abilities, and A. margalefii, A. pacificum, A. affine, A. fraterculus, and A. mediterraneum (30.0–33.0 μm) lack mixotrophic abilities. Therefore, size does not affect the presence or absence of mixotrophy in Alexandrium.
The maximum swimming speeds of the Alexandrium species with mixotrophic abilities (i.e., 160–340 μm s−1) overlapped with those lacking mixotrophic abilities (i.e., 180–680 μm s−1) (Fig. 5). To capture fast-swimming prey, mixotrophic Alexandrium species are likely faster than non-mixotrophic species. However, the maximum swimming speeds of non-mixotrophic A. pacificum, A. affine, A. fraterculus, and A. mediterraneum (i.e., 410–680 μm s−1) were considerably greater than those of mixotrophic A. andersonii, A. minutum, A. ostenfeldii, A. pohangense, A. tamarense, and A. catenella (i.e., 160–340 μm s−1). Thus, swimming speed may not affect the presence of a mixotrophy in Alexandrium.
Among the Alexandrium species tested here, every species except A. tamutum showed lytic and / or immobilizing effects on algal and ciliate prey. Prior to this study, A. catenella, A. tamarense, A. pohangense, A. pseudogonyaulax, A. ostenfeldii, and A. andersonii were known to lyse and / or immobilize algal and ciliate species (Arzul et al. 1999, Tillmann and John 2002, Tillmann et al. 2007, Tillmann and Hansen 2009, Blossom et al. 2012, Lim et al. 2015, Lee et al. 2016). The results presented here add A. insuetum, A. mediterraneum, A. pacificum, and A. margalefii to the group of Alexandrium species that can lyse and / or immobilize algal and ciliate prey. Interestingly, A. insuetum, A. mediterraneum, A. pacificum, and A. margalefii lack mixotrophic abilities, whereas A. catenella, A. tamarense, A. pohangense, A. pseudogonyaulax, A. ostenfeldii, and A. andersonii are mixotrophic. Thus, it appears A. insuetum, A. mediterraneum, A. pacificum, and A. margalefii may excrete materials that lyse and / or immobilize algal and ciliate species, even though they do not feed on them. These materials may be excreted to kill algal species competing for nutrients. The nature of the excreted materials needs to be explored further. Interestingly, A. tamutum did not lyse or immobilize algal and ciliate species, but it is known to lyse the heterotrophic dinoflagellate Polykrikos kofoidii (Kang et al. 2018). Therefore, A. tamutum excretes lysing or immobilizing materials, but the algae and ciliate appear to be tolerant of these substances.
In conclusion, the number of Alexandrium species with mixotrophic abilities is similar to the number of Alexandrium species lacking mixotrophic abilities. However, the ecological roles of mixotrophic Alexandrium species are much different from non-mixotrophic Alexandrium species. Therefore, it is important to explore the presence or absence of mixotrophic abilities in different dinoflagellate genera.
ACKNOWLEDGEMENTS
This research was supported by the Useful Dinoflagellate program of Korea Institute of Marine Science and Technology Promotion (KIMST) funded by the Ministry of Oceans and Fisheries (MOF) and the National Research Foundation (NRF) funded by the Ministry of Science and ICT (NRF-2015M1A5A1041806; NRF-2017R1E1A1A01074419) award to HJJ.
