Endoplura jejuensis sp. nov. and Endoplura koreana sp. nov. (Ralfsiales, Phaeophyceae) from Korea based on molecular and morphological analyses
Article information
Abstract
The crustose brown algal genus Endoplura has been known as a monotypic genus characterized by its intercalary plurangial reproductive structures composed of 2–4 separate parallel filaments terminated by 2–5 sterile cells and by containing several to many chloroplasts per cell. In this study, Endoplura jejuensis sp. nov. and E. koreana sp. nov. from Korea are newly described based on molecular and morphological analyses. Our phylogenetic analyses of the rbcL gene reveal that E. jejuensis sp. nov. and E. koreana sp. nov. are placed in the same clade with “E. aurea” from Japan with a strong bootstrap supporting value. E. jejuensis is characterized by small and light to dark brown crustose thalli of less than 1 cm diameter, tufts of hairs arising from the basal disc, plurangia composed mostly of two separate parallel reproductive filaments terminated by 2–4 sterile cells, and sessile unangia each with a single paraphysis. E. koreana is distinguished by olive or yellowish-brown crustose thalli of up to 3 cm diameter, tufts of hairs arising from the basal disc, and apical parts of erect filaments, plurangia with 2–5 separate reproductive filaments terminated by 2–8 sterile cells, and sessile unangia with 1–2 paraphyses. Our studies also show that “E. aurea” specimens from Japan may be recognized to be a different species from other Endoplura species.
INTRODUCTION
The genus Endoplura was originally described by Hollenberg (1969) and up to now includes only one species, Endoplura aurea Hollenberg (type locality: Orange County, CA, USA). A crustose brown algal genus, Endoplura, was distinguished from other genera of Ralfsiaceae Farlow by its intercalary plurangial reproductive structures composed of 2–4 separate parallel filaments terminated by 2–5 sterile cells and by containing several to many chloroplasts per cell (Hollenberg 1969, Abbott and Hollenberg 1976, Tanaka and Chihara 1981, Kaehler 1994). E. aurea was originally described as the generitype of Endoplura based on its vegetative and reproductive morphology (Hollenberg 1969). E. aurea has been reported in Mexico, Japan, and Hong Kong (Tanaka and Chihara 1981, Kaehler 1994, Guiry and Guiry 2021). Lim et al. (2007) identified it from Inubouzaki, Chiba, Japan, based on vegetative morphology and molecular phylogenetic analyses of the plastid ribulose-1,5-bisphosphate carboxylase large subunit (rbcL) gene. However, Endoplura has not yet been reported from Korea.
Most crustose brown algae from Korea have been known as species of Ralfsia, and Neoralfsia based on their morphology (Lee and Kang 1986, 2001, Keum 2010). However, it is difficult to correctly identify crustose species in the order Ralfsiales based on traditional morpho-anatomical characteristics because they lack robust taxonomic characters, especially for vegetative samples (Parente and Saunders 2019). Recently, plastid rbcL and mitochondrial cytochrome c oxidase 1 (COI-5P) gene sequences provide a species-level resolution to the phylogenetic classification of crustose brown algae (Lim et al. 2007, Poong et al. 2013, 2017, León-Álvarez et al. 2014a, 2014b, Silberfeld et al. 2014, Parente and Saunders 2019, Parente et al. 2021).
We collected several unidentified samples of crustose brown algae during intertidal surveys along the Korean coastal shores and recognized them as two novel species of Endoplura based on both molecular and morphological analyses. In this study, we characterize them morphologically and examine their phylogenetic relationships by rbcL and COI-5P gene sequences.
MATERIALS AND METHODS
DNA extraction and amplification
Samples were collected from intertidal areas along the western and southern coastal shores of Korea (Supplementary Table S1). Genomic DNA was extracted with the NucleoSpin Plant II Kit (Macherey-Nagel, Düren, Germany), following the manufacturer’s protocol. The extracted DNA was stored at −20°C and used to amplify the rbcL and COI-5P genes. All polymerase chain reactions (PCRs) were performed using the HelixAmp Ready-2x-Go premix (NanoHelix Co., Ltd., Daejeon, Korea) following the manufacturer’s protocol. The plastid rbcL gene was amplified and sequenced in two reactions using primer pairs NDrbcL2-DRL1R and DRL2F-R3A (Kogame et al. 1999, Hwang et al. 2005). The mitochondrial COI-5P gene was amplified and sequenced using GWSFn and GWSRx (Saunders and McDevit 2012). The PCR conditions for the rbcL gene were as described by Lee and Bae (2002) with some modifications: initial denaturation at 94°C for 4 min; 35 cycles of denaturation at 94°C for 30 s, annealing at 40°C for 30 s, and extension at 72°C for 1 min; final extension at 72°C for 10 min and storage at 4°C. PCR for the COI-5P gene was performed as described by Saunders and McDevit (2012). The additional rbcL and COI-5P sequences used in the phylogenetic analysis were selected from GenBank (Supplementary Table S1).
Sequence analyses
For phylogenetic analyses, sequence datasets were built using previously published sequences (Supplementary Table S1), as well as eight sequences (rbcL and COI-5P, respectively) newly generated in this study. The 20 rbcL and 19 COI-5P datasets were aligned using ClustalW (Thompson et al. 1994). Sargassum muticum (Yendo) Fensholt and Tilopteris mertensii (Turner) Kützing were selected as outgroups for rbcL analyses while Cladostephus spongiosus (Hudson) C. Agardh and Syringoderma phinneyi E. C. Henry and D. G. Müller for COI-5P. Before performing the phylogenetic analyses, both the best model of nucleotide and the best combination of partitions were computed using PartitionFinder 2.1.1 (Lanfear et al. 2017). Maximum-likelihood (ML) analysis was estimated by the General Time-Reversible (GTR) + Γ + I model with 1,000 bootstrap (BS) replications using RAxMLGUI v1.5 (Silvestro and Michalak 2012). Bayesian inference was performed using MrBayes 3.2.6 (Huelsenbeck and Ronquist 2001, Ronquist and Huelsenbeck 2003). Markov chain Monte Carlo runs were conducted for 2,000,000 generations, each with one cold chain and three heated chains, using the GTR + Γ + I evolutionary model and sampling and printing every 1,000 generations. Summary trees were generated using a burn-in value of 25%. Phylogenetic trees inferred from rbcL and COI-5P sequence datasets (Supplementary Table S1) were constructed to delimit the species and generic boundaries within the Ralfsiaceae. The phylogenetic trees were constructed using the ML method in RAxML and MrBayes and expressed using FigTree ver. 1.4.0 (Rambaut 2012).
Morphological analyses
To document their external morphology related to characters, including color, outline, and surface morphology, the fresh samples were photographed with a waterproof digital camera (Nikon COOLPIX AW100; Nikon Corp., Tokyo, Japan). Vouchers were air-dried and preserved in silica gel for molecular and internal morphological analyses. The samples were detached from the substrate using a single-edged blade before the analysis of morphological characters. Squashed and microtome-sectioned preparations were made for each sample. For the microtome-sectioned preparations, samples were embedded in a matrix (OCT; CellPath Ltd., Newtown, Wales, UK) and sectioned (8–10 μm thickness) using a freezing microtome (Shandon Cryotome FSE; Thermo Shandon Ltd., Loughborough, UK). Sectioned and squashed samples were stained with a 1 : 1 mixture of aqueous aniline blue and acetic acid. Sections were mounted in 50% corn syrup and photographed with a DP-71 camera (Olympus, Tokyo, Japan) mounted on a BX-51TRF microscope (Olympus). Digitized images were edited for clarity using the Adobe Photoshop software ver. 6.1 (Adobe Systems Inc., San Jose, CA, USA). Representative voucher specimens examined in this study were deposited in the herbarium of Chosun University (CUK) and the Marine Biodiversity Institute of Korea (MABIK), Korea.
RESULTS
Phylogenetic analyses
We obtained sequence data from the rbcL (1,275 bp) and COI-5P (647 bp) loci from a sample newly collected from Korea (Supplementary Table S1). Phylogenetic analyses inferred from the rbcL sequences indicated that our Endoplura specimens from Korea were recognized as two distinct new species, E. jejuensis sp. nov. and E. koreana sp. nov., within the Endoplura clade in fully supported clades (100 BS / 1.0 posterior probabilities) (Fig. 1). The COI-5P phylogenetic tree (Supplementary Fig. S1) was largely congruent to the rbcL phylogeny. Endoplura was also nested as a distinct genus within the family Ralfsiaceae. The sequence divergence for rbcL between “E. aurea” from Japan and E. jejuensis sp. nov. was 8.3%, while the divergence between “E. aurea” from Japan and E. koreana sp. nov. was 8.7–8.8%. Sequence divergences between E. jejuensis sp. nov. and E. koreana sp. nov. were 1.9–2.0% for rbcL and 11.3–11.5% for COI-5P gene.
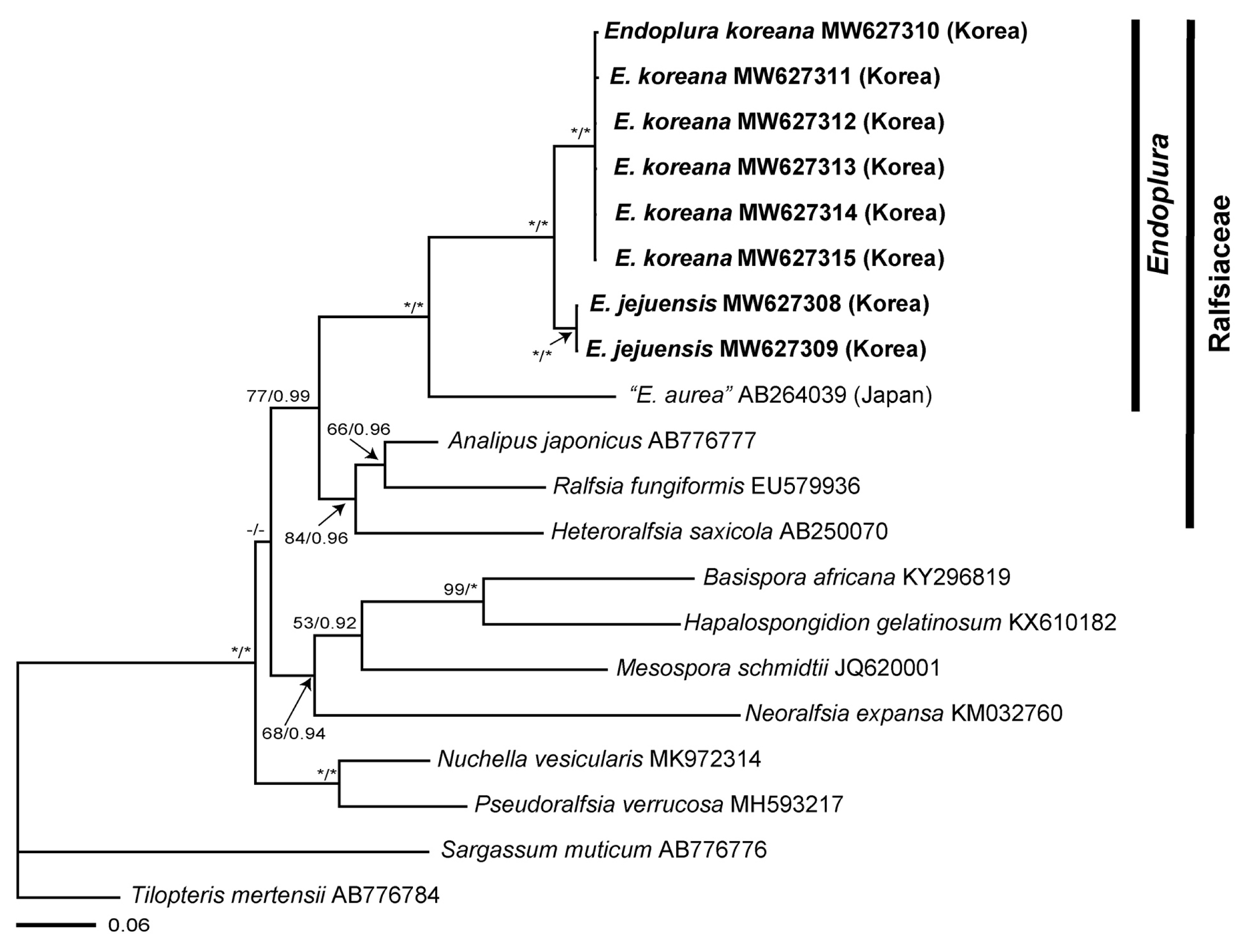
Maximum likelihood (RAxML and Bayesian inference [BI]) phylogeny of Ralfsiales inferred from gene region rbcL (1,275 bp). The value above branches = Bayesian posterior probabilities (BPP) >0.75, maximum likelihood bootstrap (BS) values in % >50. Values lower than BPP 0.75 or BS 50 are indicated by hyphens (−). Values of BPP 1.00 or BS 100 are indicated by asterisks (*). The GenBank accession numbers of each sequence data set are shown in Supplementary Table S1. Species analyzed in this study are marked in bold font. Branch lengths are proportional to substitution rate.
Morphological observations
Endoplura Hollenberg 1969 바위납작말속 (국명신칭)
Endoplura jejuensis A. O. Oteng’o, T. O. Cho & B. Y. Won sp. nov. (Fig. 2)

Endoplura jejuensis sp. nov. (A) Thalli with light to dark brown and almost circular crusts on a pebble (CUK19600). (B) Radial section view of crustose vegetative thallus showing upwardly curved erect filaments arising from indistinct hypothallial layer (CUK19630A). (C) Hairs (arrow-head) developed from the basal disc. (D) Radial section view of crustose reproductive thallus with sorus bearing plurangial reproductive filaments (Pl) on vegetative filaments (Ve) (CUK19600). (E) Plurangial reproductive structures with two separate parallel reproductive filaments (arrow) terminated by 2–4 sterile cells (arrowhead) (CUK19631A). (F) Radial section view of crustose reproductive thallus with sorus bearing unangia (CUK20757C). (G) Unangium sessile with a paraphysis (CUK20756C). Scale bars represent: A, 1 cm; B, 100 μm; C, D, F & G, 50 μm; E, 20 μm.
Description
Thalli are epilithic crusts, light to dark brown (shiny brown when dry), almost circular in outline at all stages but often confluent, with inconspicuous growth lines and ridges, less than 1 cm across, 63–317 μm thick, firmly attached to the substratum, sometimes with a rust-red undersurface, and lacking rhizoids (Fig. 2A). Vegetative erect filaments are infrequently branched, straight or upwardly curved, tapering towards the surface, firmly adjoined with one another to form pseudoparenchymatous tissue, and arise from an indistinct hypothallial layer in which cells are 3–9 μm wide and have a width to length ratio of 1 : 1.8–3 (Fig. 2B). Cells of the erect filaments are 4–20 μm long and 3–11 μm wide and contain granular storage substances. Chloroplasts are disc-shaped to irregularly discoid, several to many per cell. Tufts of hairs are sparse and arise from the basal disc (Fig. 2C). Reproductive portions of plurangial and unangial sori form elevated and irregularly confluent areas on different thalli (Fig. 2D & F). Plurangial reproductive structures are 31–98 μm long and composed mostly of two to four separate parallel reproductive filaments with 2–4 terminal sterile cells (Fig. 2E). Unangial reproductive structures are 61–116 μm long and 10–37 μm wide, oblong to obovoid, sessile, on terminal erect filaments; each sporangium accompanied by a single paraphysis (Fig. 2G). Paraphyses are present, each consisting of 7–12 cells, 73–155 μm long, and 3–14 μm wide.
Holotype
MABIK AL00080969, May 1, 2019, coll. by A. O. Oteng’o & T. O. Cho in National Marine Biodiversity Institute of Korea.
Type locality
Seongsan Ilchul Peak, Seongsan-eup, Seogwipo-si, Jeju Island, Korea (33°27′38.08″ N, 126°56′ 05.73″ E).
Isotypes
CUK19592C, CUK19607A, CUK19600, and CUK19631A in CUK herbarium, Korea.
Etymology
The specific epithet, jejuensis, refers to the geographical location of Jeju Island where the alga grows naturally.
Korean name
제주바위납작말 (국명신칭)
DNA sequences of type specimens
For the holotype, MW627308 (rbcL) and MW627316 (COI-5P).
Paratypes
CUK20756C, CUK20757B&C, CUK20759C&E, CUK20761A&B (Aug 17, 2020, Seongsan Ilchul Peak, Seongsan-eup, Seogwipo-si, Jeju Island, Korea).
Distribution and habitat
This species was confirmed on Jeju Island, Korea. It grows on hard substrates such as pebbles and rocks in the intertidal zone.
Endoplura koreana A. O. Oteng’o, T. O. Cho & B. Y. Won sp. nov. (Fig. 3)
Endoplura koreana sp. nov. (A) Thalli with brown and circular to irregular crusts with lighter margins on a pebble (CUK20628). (B) Radial section view of crustose vegetative thallus showing upwardly curved erect filaments arising from indistinct hypothallial layer (CUK19175). (C) Hair (arrow) developed from the basal disc and hair (arrowhead) developed from apical cell of erect filaments. (D) Radial section view of crustose re-productive thallus with sorus bearing plurangial reproductive filaments (Pl) on vegetative filaments (Ve) (CUK19175). (E) Plurangial reproductive structures with two to four separate parallel reproductive filaments (arrow) terminated by 2–5 sterile cells (arrowhead) (CUK20644). (F) Radial section view of crustose reproductive thallus with sorus bearing unangia (CUK20669). (G) Unangium sessile with one (right) or two (left) paraphyses (CUK20628). Scale bars represent: A, 1 cm; B & D, 100 μm; C & E–G, 50 μm.
Description
Thalli are epilithic crusts, olive or yellowish-brown to brown (shiny olive to dark brown when dry), irregular or circular in outline and often confluent, with or without lighter margins, mostly with growth lines and ridges, up to 3 cm across, 60–314 μm thick, firmly attached to the substratum, with patches of a rust-red undersurface, and without rhizoids (Fig. 3A). Vegetative erect filaments are sparsely branched, straight or upwardly curved, tapering towards the surface, firmly adjoined with one another to form pseudoparenchymatous tissue, and arise from an indistinct hypothallial layer in which cells are 3–11 μm wide and have a width to length ratio of 1 : 1.2–3 (Fig. 3B). Cells of the erect filaments are 4–20 μm long and 4–12 μm wide and contain granular storage substances. Chloroplasts are disc-shaped to irregularly discoid, several to many per cell. Phaeophycean hairs form either singly or in tufts and arise from the basal disc and apical parts of erect filaments (Fig. 3C). Reproductive portions of plurangial and unangial sori form the elevated, extensive, and irregularly confluent areas on the same or different thalli (Fig. 3D & F). Plurangial reproductive structures are 37–105 μm long, composed of 2–4 (5) separate parallel reproductive filaments terminated by 2–8 sterile cells (Fig. 3E). Unangial reproductive structures are 50–158 μm long and 10–37 μm wide, oblong to obovoid to ellipsoid, sessile, present on terminal erect filaments, and accompanied by 1–2 paraphyses (Fig. 3G). Paraphyses are present, 6–12 cells, 70–178 μm long, and 4–16 μm wide.
Holotype
MABIK AL00080968, Aug 12, 2020, coll. by A. O. Oteng’o & T. O. Cho deposited in National Marine Biodiversity Institute of Korea.
Type locality
Dumunjin Port, Baekryeongdo, Incheon, Korea (37°58′34.79″ N, 124°37′03.66″ E).
Isotypes
CUK20630, CUK20635, CUK20640, and CUK20644 in CUK herbarium, Korea.
Etymology
The specific epithet, koreana, refers to the country name of Korea where the alga grows naturally.
Korean name
한국바위납작말 (국명신칭)
DNA sequences of type specimens
For the holotype, MW627314 (rbcL) and MW627322 (COI-5P).
Paratypes
CUK18755B (Mar3, 2018, Pado-ri, Sowon-myeon, Taean-gun, Chungcheongnam-do, Korea); CUK19175A&B (Oct 8, 2018, Gijang, Ilgwang-myeon, Gijang-gun, Busan, Korea); CUK19809B, CUK19814C, CUK19818, CUK19824 (Aug 1, 2019, Oeyeondo, Ocheonmyeon, Boryeong-si, Chungcheongnam-do, Korea); CUK20574 (Jul 24, 2020, Chosa-ri, Uisin-myeon, Jindo-gun, Jeollanam-do, Korea); CUK20665, CUK20667, CUK20669, CUK20674, CUK20679, CUK20680, CUK20681 (Aug 12, 2020, Gobong Port, Baekryeongdo, Incheon, Korea).
Distribution and habitat
This species was confirmed on the western coast (Baekryeongdo, Taean, and Oeyeondo) and the southern coast (Busan and Jindo), Korea. It grows on hard substrates such as pebbles, rocks, and shells of limpets in the intertidal zone.
DISCUSSION
Research combining molecular and morphological approaches has allowed the accurate identification of crustose species of Ralfsiales (Parente and Saunders 2019, Parente et al. 2021). Our molecular analyses of crustose brown algae from Korea have expanded our understanding of the phylogenetic affinities between Endoplura and related taxa of the Ralfsiaceae and helped us infer the taxonomic position of them. This approach based on rbcL and COI-5P enabled the discovery of two new species, E. jejuensis sp. nov. and E. koreana sp. nov., thus providing useful insights into algal biodiversity (Fig. 1, Supplementary Fig. S1). E. jejuensis sp. nov. and E. koreana sp. nov. are placed in the same clade with E. aurea, type species, with a strong bootstrap supporting value. E. jejuensis sp. nov. is characterized by small and light to dark brown crust thalli of less than 1 cm, tufts of hairs arising from the basal disc, plurangia terminated by 2–4 sterile cells, and unangia each with a single paraphysis. E. koreana sp. nov. is distinguished by olive or yellowish-brown crust thalli of up to 3 cm, hair tufts arising from both basal disc and apical cell of erect filaments, plurangia terminated by 2–8 sterile cells, and unangia with one to two paraphyses. E. aurea differs from our two new species in its large and thick crusts and the absence of phaeophycean hairs (Table 1).
Endoplura was first described from Orange County, California by Hollenberg (1969). It was distinguished from other genera of Ralfsiaceae by (1) intercalary plurangial reproductive structures composed of 2–4 separate filaments, (2) 2–5 sterile cells on plurangial reproductive structures, and (3) several to many chloroplasts per cell (Hollenberg 1969, Abbott and Hollenberg 1976, Tanaka and Chihara 1981, Kaehler 1994). E. aurea was originally described as the generitype of Endoplura based on its vegetative and reproductive morphology (Hollenberg 1969) and then has been reported from Japan and Hong Kong in Asia (Tanaka and Chihara 1981, Kaehler 1994, Guiry and Guiry 2021). Recently, Lim et al. (2007) identified “E. aurea” from Chiba, Japan, based on vegetative morphology and molecular data of rbcL gene. In our molecular analyses, “E. aurea” specimens from Japan is a sister group with our E. jejuensis sp. nov. and E. koreana sp. nov. from Korea. Although E. aurea specimens from California might not be included in our analyses, the clade for Endoplura is recognized as a distinct group from other genera of Ralfsiaceae (Lim et al. 2007). However, Tanaka and Chihara (1981) showed that “E. aurea” specimens from Japan differed from E. aurea specimens from California, USA in having tufts of hairs that arose from the medial part of the thallus, plurangia with three terminal sterile cells, and upwardly curved vegetative filaments. “E. aurea” specimens from Japan seems to be a distinct species and may be recognized to be a different species from other Endoplura species.
A taxonomic study about crustose Ralfsiaceae from Korea has been carried out by combining molecular and morphological approaches (Oteng’o and Won 2020). Recent taxonomic studies on crustose brown algae in Canada, Indonesia, Japan, and Mexico indicated their great potential in diversity not only at the species level but also at the genus and family levels (Lim et al. 2007, Poong et al. 2013, 2017, León-Álvarez et al. 2017, Parente and Saunders 2019, Parente et al. 2021). Because Korea has been known as a potential hotspot of seaweed diversity for the species and generic level (Yang et al. 2020), continuous surveys about crustose brown seaweeds will contribute to the expansion of our knowledge of species diversity and ecological knowledge.
SUPPLEMENTARY MATERIALS
Supplementary Table S1. Taxa included in the phylogenetic analyses, with voucher / strain, collection locality and / or date, GenBank accession numbers for rbcL and cox1/COI-5P, and references (https://www.e-algae.org).
Supplementary Fig. S1. Maximum likelihood (RAxML and Bayesian inference) phylogeny of Ralfsiales based on COI-5P gene sequences (https://www.e-algae.org).
ACKNOWLEDGEMENTS
This study was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2019R1F1A1060346 and 2021R1I1A2059577), bythe Ministry of Ocean and Fisheris (MarineBiotics Project, 20210469), and by the National Marine Biodiversity Institute of Korea (2021M01100) to Tae Oh Cho. This research was also supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2021R1I1A1A01051909) to Boo Yeon Won.
Notes
The authors declare that they have no potential conflicts of interest.

