Unveiling mesophotic diversity in Hawai‘i: two new species in the genera Halopeltis and Leptofauchea (Rhodymeniales, Rhodophyta)
Article information
Abstract
Two genera of the Rhodymeniales, Halopeltis and Leptofauchea, are here reported for the first time from the Hawaiian Islands and represent the deepest records for both genera. Molecular phylogenetic analyses of cytochrome oxidase subunit I (COI), rbcL, and large subunit ribosomal DNA (LSU) sequences for Hawaiian specimens of Leptofauchea revealed one well-supported clade of Hawaiian specimens and three additional lineages. One of these clades is described here as Leptofauchea huawelau sp. nov., and is thus far known only from mesophotic depths at Penguin Bank in the Main Hawaiian Islands. L. huawelau sp. nov. is up to 21 cm, and is the largest known species. An additional lineage identified in the LSU and rbcL analyses corresponds to the recently described L. lucida from Western Australia, and is a new record for Hawai‘i. Hawaiian Halopeltis formed a well-supported clade along with H. adnata from Korea, the recently described H. tanakae from mesophotic depths in Japan, and H. willisii from North Carolina, and is here described as Halopeltis nuahilihilia sp. nov. H. nuahilihilia sp. nov. has a distinctive morphology of narrow vegetative axes that harbor constrictions along their length. The current distribution of H. nuahilihilia includes mesophotic depths around W. Maui, W. Moloka‘i, and the island of Hawai‘i in the Main Hawaiian Islands. Few reproductive characters were observed because of the small number of specimens available; however, both species are distinct based on phylogeny and morphology. These descriptions further emphasize the Hawaiian mesophotic zone as a location harboring many undescribed species of marine macroalgae.
INTRODUCTION
The red algal order Rhodymeniales is diverse in species richness and morphological complexity, and includes six families: Champiaceae, Faucheaceae, Fryeellaceae, Hymenocladiaceae, Lomentariaceae, and Rhodymeniaceae (Filloramo and Saunders 2016, Guiry and Guiry 2022). The order has a complicated taxonomic history, including the addition or removal of multiple families, beginning shortly after its establishment by Schmitz (Schmitz 1889, Le Gall et al. 2008). Recent studies have clarified phylogenetic relationships in the order and have resulted in new taxonomic additions at multiple levels; for example, the proposal of the families Fryeellacae and Hymenocladiacae, establishment of the genera Neogastroclonium L. Le Gall, Dalen and G. W. Saunders and Pseudohalopeltis G. W. Saunders, resurrection of the genus Halopeltis J. Agardh, and proposals for a number of new species (Gavio and Fredericq 2005, Afonso-Carrillo et al. 2006, Dalen and Saunders 2007, Le Gall et al. 2008, Schneider and Lane 2008, Rodríguez-Prieto and de Clerck 2009, Saunders and McDonald 2010, Schneider et al. 2012, Filloramo and Saunders 2015, Santiago et al. 2016).
The Rhodymeniales harbors substantial cryptic diversity (Saunders et al. 2006, Lozada-Troche and Ballantine 2010), with confusion existing even between non-sister families such as the Rhodymeniaceae and Faucheaceae (Le Gall et al. 2008). The Faucheaceae is the third largest family within the order and is characterized by three-celled carpogonial branches, terminal and cruciate tetrasporangia that are associated with adventitious cortical growth during nemathecial development, and well-developed tela arachnoidea (Saunders et al. 1999, Le Gall et al. 2008). In contrast, the Rhodymeniaceae (the largest family of the Rhodymeniales) is characterized by typically having four-celled carpogonial branches, mostly intercalary and cruciate tetrasporangia that develop in sori rather than in nemathecia, and tela arachnoidea either present or absent (Le Gall et al. 2008, Saunders and MacDonald 2010). The two families are nearly impossible to consistently tell apart at the gross morphological level because they can have very similar coloration, branching patterns, and habits.
Mesophotic coral ecosystems (MCEs) host substantial biodiversity, much of it unique, and many species have been recently described from this habitat (e.g., Ballantine et al. 2017, Schneider et al. 2019, Paiano et al. 2020, Cabrera et al. 2022). The depth range for MCEs differs based on locality but is typically defined as 30 to >150 m in tropical and subtropical areas (Lesser et al. 2009, Hinderstein et al. 2010, Baker et al. 2016, Pyle and Copus 2019). MCEs host a high level of organismal diversity (Rooney et al. 2010, Harris et al. 2013) and high levels of endemism (Kane et al. 2014, Baker et al. 2016, Pyle et al. 2016, Kosaki et al. 2017). This holds true for Hawaiian MCEs, where endemism levels are also high due to the isolation of the archipelago (Grigg 1988), and several new-to-science mesophotic algal species have recently been described (e.g., Spalding et al. 2016, Sherwood et al. 2020, Cabrera et al. 2022). Macroalgae are abundant in Hawaiian MCEs (Spalding et al. 2019), and algal beds have been found as deep as 160 m (Spalding 2012). Influxes of cold water in the Hawaiian mesophotic provide suitable habitat for algae typically found in temperate zones (Abbott and Huisman 2003), resulting in certain species and genera being found only in the mesophotic and absent from the better-studied adjacent shallow waters (Cabrera et al. 2022). In the Main Hawaiian Islands, mesophotic macroalgae may also be influenced by nearshore anthropogenically derived nitrogen via submarine groundwater discharge (Strait et al. 2022).
In Hawai‘i, seven genera of the family Rhodymeniaceae have been previously reported (Botryocladia, Chrysymenia, Coelarthrum, Drouetia, Erythrocolon, Halichrysis, and Rhodymenia), and two from the Faucheaceae (Gloiocladia and Gloioderma) (Abbott and Littler 1969, Abbott 1999). Neither genus examined in this study (Leptofauchea and Halopeltis) has been previously documented in the Hawaiian Islands. Leptofauchea (Faucheaceae) contains 11 taxonomically accepted species, while Halopeltis (Rhodymeniaceae), a resurrected genus as of 2010 (Saunders and McDonald 2010), currently contains 10 species (Guiry and Guiry 2022). This study describes the phylogenetic relationships for Hawaiian representatives of these two genera, presents the description for one new species in each genus, and examines the biogeographic patterns for these new additions to the Hawaiian flora.
MATERIALS AND METHODS
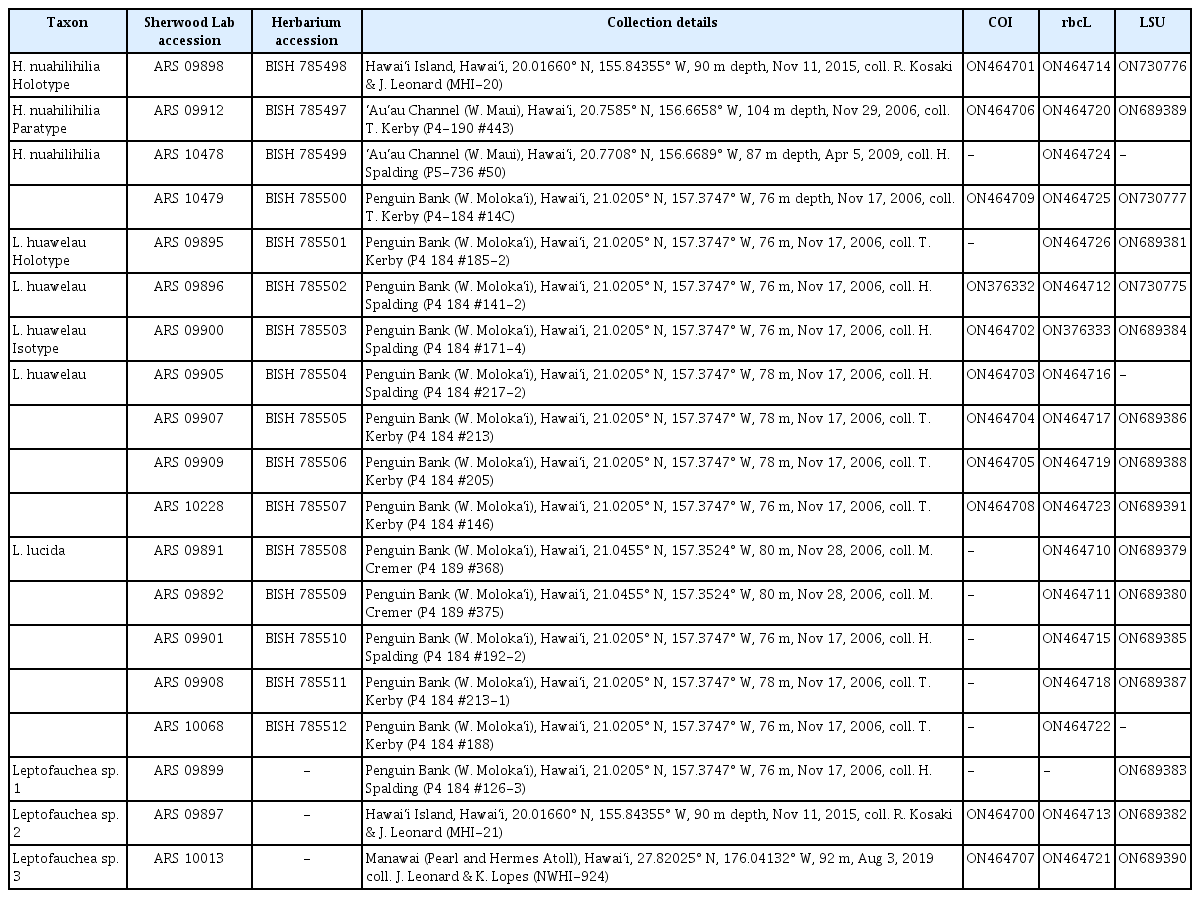
Mesophotic algal specimens were collected by technical divers or the Pisces IV and Pisces V submersibles within the Main Hawaiian Islands and the Papahānaumokuākea Marine National Monument (PMNM). Most specimens were collected in 2006 off the west coast of Moloka‘i (Penguin Bank). Two specimens were collected from the island of Hawai‘i in 2015, and one was collected from PMNM in 2019. After collection, samples were mounted as herbarium specimens and preserved in silica gel for molecular analyses. Specimen collection information is listed in Table 1.
DNA was extracted with an OMEGA E.Z.N.A Plant DNA Kit (OMEGA Biotek, Norcross, GA, USA), following the manufacturer’s protocols. Amplification of the 5′ end of the mitochondrial cytochrome c oxidase I (COI-5P) barcode was accomplished using one of two sets of primers: GazF1 and GazR1 (Saunders 2005), or GWSFn and GWSRx (Saunders and Moore 2013). The plastidial rbcL gene was amplified in three fragments: F57 and R562 (Freshwater and Rueness 1994) or F8 and R646 for the first fragment, F481 and R1150 for the second, and F765 and R1381 for the third fragment (Wang et al. 2000). For the large subunit ribosomal DNA (LSU) marker, four pairs of primers and cycling conditions were used: T16 and T24, T25 and T20, T05, and T15 (Saunders and Moore 2013), and nu28SF and nu28SR (Sherwood et al. 2010). Successful polymerase chain reaction products were submitted for sequencing by GENEWIZ (South Plainfield, NJ, USA).
Editing and aligning of sequence data was conducted in Geneious Prime with downloaded reference sequence data from GenBank and BOLD (Barcode of Life Database) (Supplementary Table 1). Sequences of the three markers were aligned with MUSCLE v. 3.8.425 plug-in (Edgar 2004) using the default settings; sequences were aligned separately for each marker to check for inconsistencies, and phylogenetic analyses were conducted on the individual marker alignments as well as a concatenated alignment of taxa for which sequences of all three markers were available. The Halopeltis alignments included 26 rbcL sequences (1,383 bp), 33 COI sequences (664 bp), and 28 LSU sequences (2,717 bp), while the Leptofauchea alignments included 26 rbcL sequences (1,362 bp), 20 COI sequences (664 bp), and 25 LSU sequences (2,764 bp). A series of taxa were used as outgroups in the Halopeltis phylogeny, as per Schneider et al. (2012), and Webervanbossea was used as the outgroup for Leptofauchea (Filloramo and Saunders 2015). Alignments were then uploaded to CIPRES (Miller et al. 2010). JModelTest2 was used to determine the best fit model of evolution (in each case GTR + Γ + I) for individual marker alignments, and PartitionFinder 2.1.1 was used for the concatenated alignment (Lanfear et al. 2017) which was run with the greedy algorithm (Lanfear et al. 2012) with unlinked branch lengths and Bayesian information criterion as the selection criterion. Maximum likelihood phylogenies were constructed using RAxML (Stamatakis 2014) with 1,000 permutations and four threads on the University of Hawai‘i High Performance Computing (HPC) cluster (https://datascience.hawaii.edu/hpc). Bayesian analyses were conducted using MrBayes 3.2.7 (Ronquist et al. 2012) on HPC under 1,000,000 generations using four chains of Metropolis-coupled Markov Chain Monte Carlo with sampling every 1,000 generations. Sequences were submitted to GenBank and are available as accessions ON376332 and ON464700–ON464709 (COI), ON376333 and ON464710–ON464726 (rbcL), and ON689379–ON 689391 and ON730775–ON730777 (LSU). Phylogenetic analyses are presented as concatenated marker trees for Leptofauchea (Fig. 1) and Halopeltis (Fig. 2), with analyses of individual markers included in Supplementary Figs S1–S6.
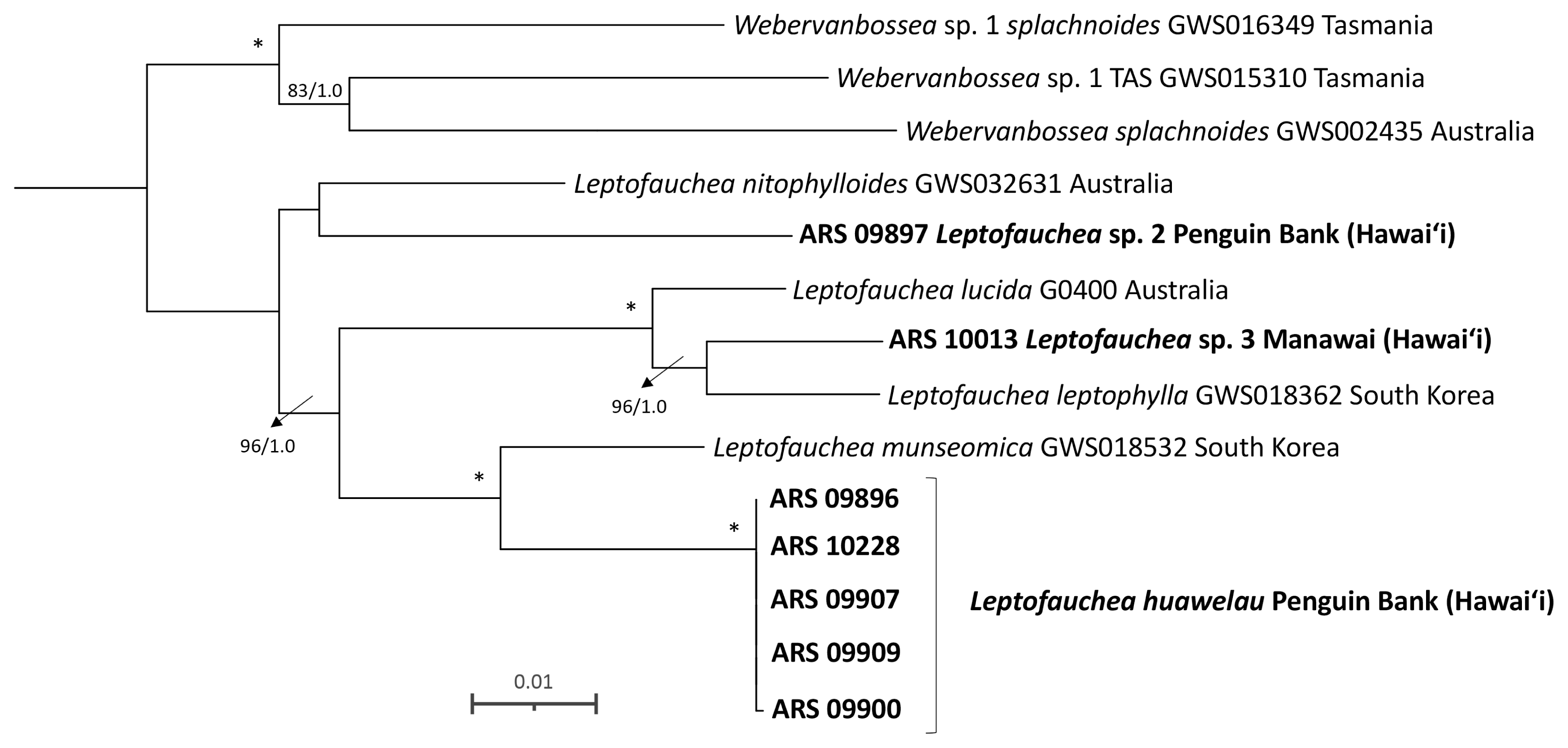
Phylogenetic tree of the genus Leptofauchea using a concatenated alignment of 5′ end of the mitochondrial cytochrome c oxidase I, large subunit ribosomal DNA, and rbcL sequences. Sequences generated in the current study are indicated in bold. Bayesian and maximum likelihood support values shown at the nodes, the first value represents the maximum likelihood bootstrap support (BP) value and the second is the Bayesian posterior probability (PP) support value. Full support is indicated with an asterisk (*) while support less than 70% BP / 0.70 PP is shown with a dash (−). Scale bar represents: substitutions per site.

Phylogenetic tree of the genus Halopeltis using a concatenated alignment of 5′ end of the mitochondrial cytochrome c oxidase I, large subunit ribosomal DNA, and rbcL sequences. Sequences generated in the current study are indicated in bold. Bayesian and maximum likelihood support values shown at the nodes, the first value represents the maximum likelihood bootstrap support (BP) value and the second is the Bayesian posterior probability (PP) support value. Full support is indicated with an asterisk (*) while support less than 70% BP / 0.70 PP is shown with a dash (−). Scale bar represents: substitutions per site.
Anatomical features were observed by mounting specimens onto microscope slides and examining with a Zeiss AxioImager A1 compound light microscope (Pleasanton, CA) with an Infinity2-1RC digital camera (Lumenera Corporation, Ottawa, ON, Canada). Slides were made by hand-sectioning material using a razor blade and hydrating in water for 5 min followed by staining for 5 min with 0.5% aniline blue before mounting in 30% Karo. Digitization of herbarium sheets was performed in the Joseph F. Rock Herbarium (HAW) with a Canon EOS 5D Mark II Digital Camera (Tokyo, Japan) mounted on an MK Direct Photo-eBox PLUS 1419. Vouchers were accessioned at the Bernice P. Bishop Museum (BISH) under accession codes BISH785497–785512.
RESULTS
Concatenated COI + rbcL + LSU phylogenetic analyses of sequences of the genus Leptofauchea demonstrated that the mesophotic Hawaiian samples are distinct from all but two previously described species (L. auricularis E. Y. Dawson and L. rhodymenioides W. R. Taylor, which could not be ruled out due to a lack of confirmed molecular data for these species) (Fig. 1). Hawaiian mesophotic Leptofauchea samples were resolved in one clade in the concatenated analysis, with two additional specimens forming distinct lineages in other parts of the tree (Leptofauchea sp. 2 and sp. 3) (Fig. 1). A fourth lineage of Hawaiian specimens in the LSU (Supplementary Fig. S3) and rbcL phylogenies (Supplementary Fig. S1) corresponded to a recently described species from Australia, L. lucida Huisman and G. W. Saunders (Filloramo and Saunders 2015, Huisman and Saunders 2020). Specimens in this clade shared the key morphological features Huisman and Saunders (2020) described for this species, including a sprawling habit, abundant secondary anastomoses between branches, and a multi-layered cortex (e.g., Supplementary Fig. S7D). A fifth lineage (Leptofauchea sp. 1) was recovered only in the LSU single-marker analysis (Supplementary Fig. S3). The clade containing most of the Hawaiian mesophotic specimens in the concatenated analysis, which is described in the next section as a new species, includes five Hawaiian specimens from Penguin Bank, MHI (ARS 09896, 09900, 09907, 09909, 10228), and belongs to a clade with L. munseomica Filloramo and G. W. Saunders, from South Korea, with full support (Filloramo and Saunders 2015).
All currently described species of Halopeltis are represented in the concatenated phylogeny and / or individual marker trees, as well as several additional lineages of undescribed species, labeled in GenBank as “GWS” collections (Fig. 2, Supplementary Figs S4–S6). The Hawaiian mesophotic samples sequenced in this study form a well-supported clade in the concatenated analysis with full support that is distinct from all other available sequences for the genus. Three MHI samples are included within this clade (ARS 09898, 09912, 10479), which belongs to a larger clade that also includes H. adnata (Okamura) G. W. Saunders and C. W. Schneider, H. tanakae Mas. Suzuki and R. Terada, and H. willisii Freshwater and G. W. Saunders with full support (Schneider et al. 2012, Suzuki and Terada 2021).
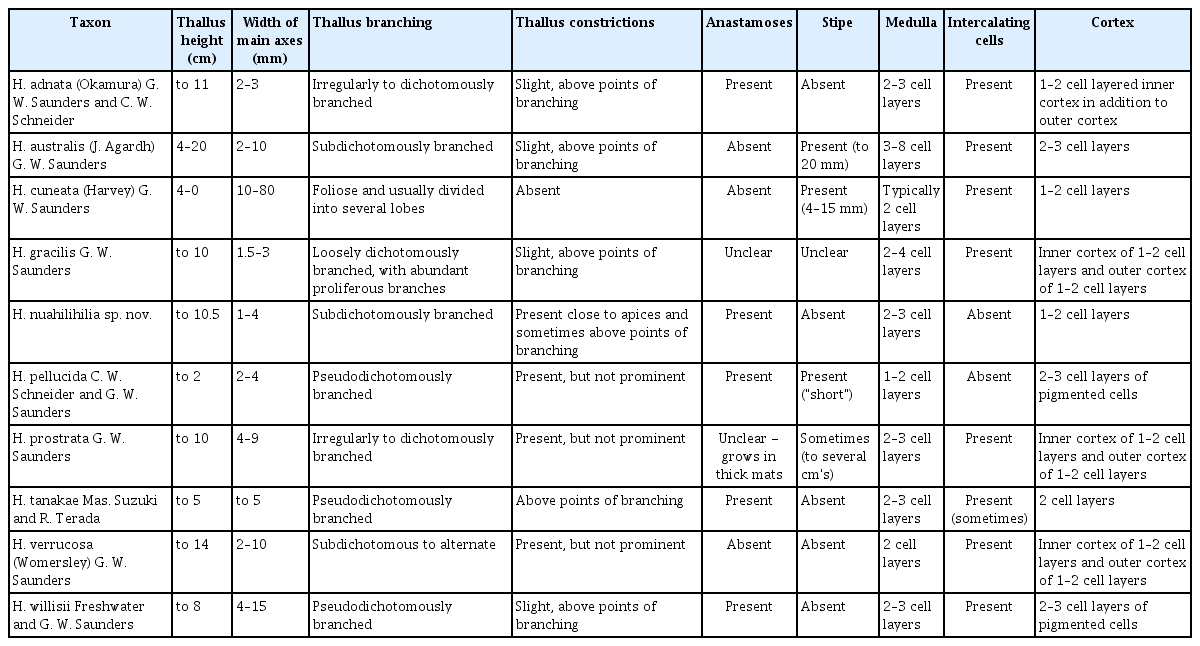
The formal descriptions for the one new species each of Leptofauchea and Halopeltis from the Hawaiian specimens are presented below, with comparisons of general thallus morphology among related species presented in Table 2 (Leptofauchea) and Table 3 (Halopeltis).
Leptofauchea huawelau E. A. Alvarado, F. P. Cabrera and A. R. Sherwood sp. nov. (Fig. 3A–I)
Morphology of Leptofauchea huawelau sp. nov. (A) Image of the herbarium sheet containing the holotype (BISH 785501, ARS 09895). (B) Close up view of the holotype (BISH 785501, ARS 09895). (C) Cross section of the stipe (BISH 785501, ARS 09895). (D) Cross section of apical portion of the thallus, showing the cortex and medulla (BISH 785501, ARS 09895). (E) Surface view of the blade showing outer cortical cells (BISH 785501, ARS 09895). (F) Cruciately divided tetraspores (BISH 785505, ARS 09907). (G) Cross section of basal portion of the thallus, showing the cortex and medulla (BISH 785501, ARS 09895). (H) Tetrasporangia are laterally produced from paraphyseal cells or terminally on short filaments (BISH 785505, ARS 09907). (I) Close up of early developmental stages of tetrasporangia (BISH 785505, ARS 09907). Scale bars represent: A & B, 5 cm; C, D & H, 100 μm; E, 25 μm; F, G & I, 50 μm.
Description
Thalli up to 21 cm in height × 18 cm in width, attached by a stipe, light pink to deep red-pink in color. Plants tall, with irregular to loosely dichotomous branching. Vegetative axes narrow, 2–4 mm broad with constrictions frequently present below or above branching points, constrictions ranging in breadth from <1–1.75 mm. Internodal lengths of axes between points of branching up to 6.4 cm. Axes flat and thin, ranging in thickness from 120–150 μm. Apices commonly spatulate, lighter in color than other sections of the thallus, occasionally attenuated. Cortical cells loosely arranged in surface view, darkly staining, with a single outer layer, cells spherical to slightly axially elongated, 7.4–17 μm in diameter. Inner cortex composed of 1–2 incomplete layers of cells, which grade into the medulla. Medulla composed of 2–3 layers of cells, irregularly arranged and visible from surface view. Medullary cells large and axially elongated, 27–81 μm in length × 18–47 μm in width, lightly staining, and thin-walled. Tetrasporangial nemathecia slightly darker than the surrounding thallus, positioned near apices, round to elliptical in shape, and elongated towards the branch apex, 1,370 × 740 μm, with 4–7-celled paraphyses. Tetrasporangia cruciate or decussate, spherical to ovoid, 7.6–29.8 μm in diameter. Gametangial reproduction not observed.
Holotype
BISH 785501 (ARS 09895, West Moloka‘i, Hawai‘i, 21.022172° N, 157.216848° W, 72 m depth, Nov 17, 2006, leg. Terry Kerby, field code P4-184 #185).
Holotype DNA accession numbers
ON464726 (rbcL), ON689381 (LSU).
Isotype
BISH 785503 (ARS 09900).
Isotype DNA accession numbers
ON464702 (COI), ON376333 (rbcL), ON689384 (LSU).
Etymology
Specific epithet derived from the Hawaiian language, pronounced “hoo-ah-veh-la-oo.” “Hua” is often translated as “fruit, egg, seed.” “Wēlau” means “tip” or “end.” Simply put, this name describes the “bulbs” at the apices of the branches that reflect its growth potential. New branches bud from these hua, creating more bulbs and continued growth. The first part of the name defines the epithet as a noun, and is thus not declinable in Latinized form.
Distribution
Mesophotic depths of Penguin Bank, Main Hawaiian Islands, Hawai‘i, USA.
Specimens examined
BISH 785501–785507 (ARS 09895, 09896, 09900, 09905, 09907, 09909, 10228).
Habit and morphology
Individuals are flattened, loosely dichotomously branched, attached by a stipe (Fig. 3A & B). Stipe 0.2–0.4 cm in length, 950–1,200 μm in width (Fig. 3A–D), with a multi-layered cortex and medulla. Dried specimens have a deep pink-red color that, in most cases, lightens to a very light pink near branch apices. Thalli with a lanky appearance, with long internodes between points of branching (Fig. 3B). Thalli thin in cross section, 120–150 μm (Fig. 3D) with large medullary cells in 2–3 layers and small pigmented cortical cells in 1–2 incomplete layers (Fig. 3G). In surface view cortical cells are loosely arranged, cortical cells small, mostly round, 7.4–17 μm in diameter (Fig. 3E). Medulla is 2–3 layers thick, with thin-walled cells, 27–81 μm long by 18–47 μm wide (Fig. 3D & G). Cruciate or decussate tetrasporangia are 7.6–29.8 μm in diameter, 11.5–26.0 μm in length (Fig. 3F). Tetrasporangia formed in nemathecia that are elongated towards branch apices, 1,370 × 740 μm in size. Nemathecial paraphyses composed of 4–7 cells (Fig. 3H & I). No gametangial reproduction has been confirmed in the specimens analyzed thus far for this species; however, all collections were made in the month of November, and it is possible that future collections representing other seasons will help to elucidate these characters.
Halopeltis nuahilihilia E. A. Alvarado, F. P. Cabrera and A. R. Sherwood sp. nov. (Fig. 4A–F)
Morphology of Halopeltis nuahilihilia sp. nov. (A) Image of the herbarium sheet of the holotype (BISH 785498, ARS 09898). (B) Image of the herbarium sheet of a paratype (BISH 785497, ARS 09912). (C) Surface view of the blade showing outer cortical cells (BISH 785498, ARS 09898). (D) Cross section of thallus, showing cortex and medulla (BISH 785498, ARS 09898). (E) Cross section of a developing spermatangial sorus (BISH 785498, ARS 09898). (F) Close up of spermatangia produced terminally on spermatangial mother cells (BISH 785498, ARS 09898). Scale bars represent: A & B, 5 cm; C, 25 μm; D, 100 μm; E & F, 50 μm.
Description
Thalli up to 10.5 cm tall and 7.3 cm wide, including anastomosed portions. Plants layered on top of each other creating a tangled appearance in situ, spreading, with multiple anastomoses, attached to substratum by haptera. Thalli deep pink-purple to medium-pink in color. Vegetative axes narrow, 1–4 mm wide, subdichotomously branched. Constrictions along axes typically close to apices and sometimes close to points of branching. Vegetative axes flat and thin, up to 190 μm in cross section. Apices attenuate, spatulate, rounded, or ovular. Outer cortex composed of 1–2 layers, inner cortical layer incomplete. Cortical cells small and darkly staining, positioned irregularly, 11–17 μm by 8.5–13 μm. Medullary cells round to slightly elongated axially, 40–117 μm long by 40–79 μm wide, medullary cells in 2–3 layers, lightly staining. Small, intercolating cells absent. One or two elongate spermatangia measuring up to 4–15 μm long by 1.4–5 μm wide produced terminally by spermatangial mother cells 8.3–14 μm wide, formed in apical sori on the branches. Female gametangial and tetrasporangial reproduction not observed.
Holotype
BISH 785498 (ARS 09898, Hawai‘i Island, Hawai‘i, 20.016600° N, 155.843550° W, 90 m depth, Nov 11, 2015, leg. Randall Kosaki and Jason Leonard, field code MHI-20).
Holotype DNA accession numbers
ON464701 (COI), ON464714 (rbcL), ON730776 (LSU).
Paratype
BISH 785497 (ARS 09912, ‘Au‘Au Channel, Maui, Hawai‘i, 20.458500° N, 156.396000° W, 104 m depth, Nov 29, 2006, leg. Terry Kerby, field code P4-190 #443).
Paratype DNA accession numbers
ON464706 (COI), ON464720 (rbcL), ON689389 (LSU).
Etymology
Specific epithet derived from the Hawaiian language, pronounced “noo-ah-hee-lee-hee-lee-ah.” “Nu‘a” describes the thick-growing nature of this alga. Additionally, “nu‘a” refers to the way this alga grows, spreading out and appearing as a single mass of different branches connecting to each other. “Hilihili” is the action of braiding or plaiting. Its use in the name honors the twisted-like appearance of this alga, and its intertwining with other algal species. Moreover, “hili”, which translates as “dark red”, appropriately reflects the color of this species.
Distribution
Mesophotic depths of Hawai‘i Island, W. Moloka‘i, and W. Maui, Hawai‘i, USA.
Specimens examined
BISH 785497-785500 (ARS 098 98, ARS 09912, ARS 10478, ARS 10479).
Habit and morphology
Specimens have a range of morphologies, from broadly branched, to less so (Fig. 4A & B). Specimens range in color from dark pink (Fig. 4A) to light pink (Fig. 4B). Apices are varied and present a variety of shapes: rounded, attenuate, and ovular. From a surface view, cortical cells are arranged loosely and irregularly, darkly staining (Fig. 4C). The outer cortex is composed of 1–2 layers with an incomplete inner layer, cells ranging in size from 11–17 μm by 8.5–13 μm (Fig. 4C & D). Medulla composed of 2–3 layers of these lightly staining cells (Fig. 4D). Medullary cells round to axially elongated, large and thin-walled, 40–117 μm long by 40–79 μm in width (Fig. 4D). Small, intercolating cells (which are reported for most species of Halopeltis) appear to be absent in the medulla of H. nuahilihilia. One or two spermatangia, 4–15 μm long by 1.4–5 μm wide in diameter, are grouped in surface sori situated in the apical parts of the branches, elongating from ovoid spermatangial mother cells 8.3–14 μm wide, that develop from the outer cortical cells (Fig. 4E & F). No other reproductive stages were observed.
DISCUSSION
Leptofauchea and Halopeltis are both new genus records for the Hawaiian Islands and contribute to the growing list of such additions to the Hawaiian flora (e.g., Huisman and Abbott 2003, Kraft et al. 2004, Sherwood and Carlile 2012, Conklin et al. 2014). Interestingly, specimens of both genera have very similar gross morphology and were collected from similar environments. Hawaiian Leptofauchea specimens were discovered growing in clumps among other macroalgal species, dispersed loosely in a sandy habitat, which resembles reports for other Leptofauchea species (Dalen and Saunders 2007). Leptofauchea, although largely reported from shallow waters, has been reported from depths greater than 30 m in other geographical regions (Gavio and Fredericq 2005, Dalen and Saunders 2007, Rodríguez-Prieto and de Clerck 2009), but Halopeltis had only been reported to 30 m (Saunders and McDonald 2010, Schneider et al. 2012) until the recent description of H. tanakae Mas. Suzuki and R. Terada from 50 m (Suzuki and Terada 2021). Thus, the samples studied here from the Hawaiian Islands are the deepest reported collections for both genera. Intriguingly, however, Leptofauchea and Halopeltis are thus far only known from the mesophotic zone in Hawai‘i and have not been documented from depths shallower than 76 m in Hawai‘i. These genera add to our knowledge of algae present in MCEs of Hawai‘i, which have a high level of endemism for many groups of organisms (Kane et al. 2014, Baker et al. 2016, Pyle et al. 2016, Kosaki et al. 2017).
Of the Hawaiian Leptofauchea lineages revealed in the phylogenetic analyses, two can be assigned taxonomic names (either from previously described species or newly named here). Three specimens of Leptofauchea collected in Hawai‘i (ARS 09897, ARS 09899, and ARS 10013) are included in the molecular phylogenetic analyses but are positioned with low-to-no support. Due to the poor phylogenetic resolution of these specimens, and the small number of specimens available per lineage, recognition of these as new species will await collection of additional specimens to include in the analyses. Analyses yielded full support for L. huawelau sp. nov., which is a member of a larger clade that includes L. munseomica. Morphologically, L. huawelau is distinct from other species based on overall thallus shape: plants are remarkably long (reaching 21 cm in height) and narrow (typically 2–4 mm), giving them a “leggy” appearance (Table 2) (Gavio and Fredericq 2005, Dalen and Saunders 2007, Rodríguez-Prieto and de Clerck 2009, Suzuki et al. 2012, Filloramo and Saunders 2015). The spatulate apices, resulting from constrictions below the apices, are similar to those reported for L. leptophylla (Suzuki et al. 2012). The second Hawaiian species corresponded to the recently described L. lucida from Western Australia (Huisman and Saunders 2020). The Hawaiian specimens of L. lucida share diagnostic characters with specimens from Australia, including the sprawling and anastomosed habit with frequent secondary anastomoses between branches and a multi-layered cortex (Huisman and Saunders 2020).
Hawaiian specimens of Halopeltis nuahilihilia sp. nov. exhibited different degrees of maturation and consequently some variation in overall morphology, which provides insight into the plasticity of these features in the species. The most distinctive differences between H. nuahilihilia and other described species are the combination of narrow vegetative axes, the presence of constrictions along these axes in the Hawaiian specimens, and the apparent lack of small, intercolating cells in the medulla (Table 3). While constrictions are present in H. pellucida (Schneider et al. 2012), H. verrucosa, and H. prostrata (Saunders and McDonald 2010), they are more prominent in the mesophotic Hawaiian specimens.
While Leptofauchea huawelau sp. nov. is thus far known only from Penguin Bank in the MHI, Hawaiian Leptofauchea specimens were collected from a broad geographical range that extends from Hawai‘i Island in the southeast of the archipelago to PMNM in the northwest. The L. lucida specimens from Hawai‘i extend the known range of this species from shallower habitats (5–37.1 m) in Western Australia (Huisman and Saunders 2020) to 80 m depth in Hawaiian MCEs. In addition, L. huawelau is sister to L. munseomica, known only from South Korea (Filloramo and Saunders 2015). Many marine algal species from Hawai‘i have been demonstrated to be most closely related to taxa from the Northwestern Pacific or Australia (Abbott and Huisman 2003, McDermid and Abbott 2006, Spalding et al. 2016, Paiano et al. 2020, Sherwood et al. 2020, Cabrera et al. 2022), which suggests an interesting biogeographic connection between the two regions, but more information on the mesophotic flora of the Pacific Islands is needed to know the true range and biogeographic processes that account for this distribution. In contrast to the broad distributional range of Leptofauchea in Hawai‘i, Hawaiian Halopeltis specimens were collected from a much narrower range, which includes only Maui to Hawai‘i Island in the MHI (the southeasternmost part of the archipelago). H. nuahilihilia sp. nov. is most closely allied with H. adnata from Korea and H. tanakae from Japan (Schneider et al. 2012, Suzuki and Terada 2021), suggesting biogeographical affinities between Hawaiian species and species from eastern Asia.
While about 80% of all coral reef habitat is included in MCEs (Pyle and Copus 2019), most marine biodiversity studies focus on shallow reefs, yielding a biased impression of marine diversity, function, and ecology. Uncovering hidden diversity in MCEs can help understand evolutionary and biogeographic patterns by adding to the current body of knowledge. Similarly, having adequate representation of mesophotic species in phylogenetic analyses can illuminate the links between shallow and deep-water species. However, there is still much to be done to categorize macroalgal diversity of mesophotic habitats. Multiple marine macroalgal species have been newly described from the Hawaiian mesophotic (e.g., Spalding et al. 2016, Sherwood et al. 2019, 2020, Paiano et al. 2020, Cabrera et al. 2022) and these species represent just a small fraction of the diversity that has yet to be categorized (Sherwood et al. 2010). The two new red algal genera reported here from the Hawaiian mesophotic, Leptofauchea and Halopeltis, demonstrate the need for a better understanding of the biodiversity of deep reefs.
ACKNOWLEDGEMENTS
We gratefully acknowledge the NOAA Papahānaumokuākea Native Hawaiian Cultural Working Group for their invaluable contributions in developing the specific epithets for the Hawaiian species of Halopeltis and Leptofauchea. G. McFall, S. Matadobra, B. Hauk, T. Efird, A. Fukunaga, D. Wagner, and H. Owen assisted with diving and small boat operations. Our gratitude goes to Terry Kerby and the Hawai‘i Undersea Research Laboratory (HURL) Pisces IV and V submersible and RCV-150 pilots and crew, as well as the crew of the R/V Ka’imikai-o-Kanaloa, for access to mesophotic habitats around the Main Hawaiian Islands. Field work and specimen collections in the Papahānaumokuākea Marine National Monument were authorized under Papahānaumokuākea Marine National Monument research permit PMNM-2019-001 issued to R. Kosaki. This work was supported by the U.S. National Science Foundation (DEB-1754117), the U.S. National Fish and Wildlife Foundation (NFWF 0810.18.059023), the National Oceanic and Atmospheric Administration (NOAA) Papahānaumokuākea Marine National Monument, NOAA Coastal Ocean Program (NA07NOS4780187 and NA07NOS478190 to the University of Hawai‘i), NOAA’s Undersea Research Program and Coral Reef Conservation Program through the Hawai‘i Undersea Research Laboratory (NA09OAR4300219 and NA05OAR4301108), and NOAA’s Office of Ocean Exploration. The scientific views and conclusions, as well as any views or opinions expressed herein, are those of the authors and do not necessarily reflect the views of the above organizations.
Abbreviations
BOLD
Barcode of Life Database
COI
cytochrome oxidase subunit I
HPC
High Performance Computing
LSU
large subunit ribosomal DNA
MCE
Mesophotic Coral Ecosystem
MHI
Main Hawaiian Islands
NOAA
US National Oceanographic and Atmospheric Administration
NWHI
Northwestern Hawaiian Islands
PMNM
Papahānaumokuākea Marine National Monument
rbcL
ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit gene
Notes
The authors declare that they have no potential conflicts of interest.
SUPPLEMENTARY MATERIALS
Voucher and GenBank accession information for sequences used in phylogenetic analyses in the current study (https://www.e-algae.org).
Phylogenetic tree of the genus Leptofauchea based on 5′ end of the mitochondrial cytochrome c oxidase I (COI-5P) sequences (https://www.e-algae.org).
Phylogenetic tree of the genus Leptofauchea based on rbcL sequences (https://www.e-algae.org).
Phylogenetic tree of the genus Leptofauchea based on large subunit ribosomal DNA (LSU) sequences (https://www.e-algae.org).
Phylogenetic tree of the genus Halopeltis based on 5′ end of the mitochondrial cytochrome c oxidase I (COI-5P) sequences (https://www.e-algae.org).
Phylogenetic tree of the genus Halopeltis based on rbcL sequences (https://www.e-algae.org).
Phylogenetic tree of the genus Halopeltis based on large subunit ribosomal DNA (LSU) sequences (https://www.e-algae.org).
Morphology of Leptofauchea lucida (https://www.e-algae.org).