Transcriptome analysis revealed regulatory mechanisms of light and culture density on free-living sporangial filaments of Neopyropia yezoensis (Rhodophyta)
Article information
Abstract
Previous research indicated that free-living sporangial filament keep hollow morph under high-culture density and form bipartite cells under low-culture density, while the following conchospore release was inhibited by high light. Here, we further explored the molecular bases of these affects caused by light and culture density using a transcriptome analysis. Many differentially expressed genes (DEGs) related to carbon dioxide concentration and fixation, photosynthesis, chlorophyll synthesis and nitrogen absorption were upregulated under high-light conditions compared with low-light conditions, indicating the molecular basis of rapid vegetative growth under the former. The stress response- and ion transport-related DEGs, as well as the gene encoding the vacuole formation–brefeldin A-inhibited guanine nucleotide exchange protein (BIG, py05721), were highly expressed under high-density conditions, indicating the molecular basis of the hollow morph of free-living sporangial filaments under high-culture density conditions. Additionally, the brefeldin A treatment indicated that the hollow morph was directly influenced by vacuole formation-related vesicle traffic. Others DEGs related to cell wall components, zinc-finger proteins, ASPO1527, cell cycle and cytoskeleton were highly expressed in the low density with low-light group, which might be related to the formation and release of conchospores. These results provide a deeper understanding of sporangial filaments in Neopyropia yezoensis and related species.
INTRODUCTION
Neopyropia yezoensis is an economically important red algae and widely cultivated in East Asian countries, including China, Japan and Korea (Kim et al. 2017). The annual yield and market of Neopyropia / Neoporphyra reaches 110,000 tons and 0.95 billion dollars, respectively (Kim et al. 2017). The life cycle of Neopyropia yezoensis is complex (Supplementary Fig. S1), which is mainly composed of diploid sporophyte generation (including conchocelis and sporangial filament) and haploid gametophyte generation (thallus). The alternation between generations is connected by three types of spores, such as conchospore mediate sporophyte (sporangial filament) to gametophyte (thallus), zygotospore mediate gametophyte (mature thallus) to sporophyte (conchocelis), and archeospore mediate the asexual reproduction of gametophyte (Supplementary Fig. S1) (Wang et al. 2020b). In the large-scale artificial cultivation of Neopyropia / Neoporphyra, conchospores are the important sources of “seeds”, which are usually collected and adhered on nets used for the subsequent cultivation in intertidal zone (Wang et al. 2020b).
Sporangial filaments represent late-stage sporophytes in the life cycle of N. yezoensis and can directly release conchospores after maturation. Hence, sporangial filaments are very important in seedling collection for the large-scale artificial breeding of Neopyropia / Neoporphyra. Prior to conchospore release, the appearance of massive bipartite cells in the sporangial filaments is an important sign (Supplementary Fig. S1) (Chen 1980, He et al. 2021b). The traditional seedling collection is formed on the bases of the “shell inoculation” method, in which sporangial filaments form and mature in shell, but this is a time-consuming and labor-intensive process (Li et al. 2011). Using free-living sporangial filaments for seedling development would be more economical if conchospore release could be accurately regulated. Although many studies have explored the optimal conditions for the formation, maturation and conchospore release of free-living sporangial filaments (Chen 1980, Sun and Zeng 1996, Yang and He 2004, Li et al. 2011, López-Vivas et al. 2015), to date, the use of seedling collections from free-living sporangial filaments has not been popularized in large-scale production. The main reason is still the inability to accurately control the massive free-living sporangial filaments’ release of conchospores.
Recently, we reported the periodic morphological changes between hollow cells and bipartite cells, as well as the regulation of conchospore release of free-living sporangial filaments by culture density and light intensity (He et al. 2021b). We found that high-culture density (5,000–10,000 fragments mL−1) made sporangial filaments remain a hollow morph, in which vacuole expansion without bipartite cells formation and conchospore release has been observed (He et al. 2021b). Moreover, the bipartite cells themselves can return to vegetative growth, especially under high light (60–100 μmol photons m−2 s−1) conditions, thereby the subsequent conchospore release is inhibited (He et al. 2021b). In this paper, we established up three N. yezoensis groups for a transcriptome analysis to further explore the molecular basis of the effects of light and culture density on free-living sporangial filaments. The low light with low density (LD) condition was set as control under which bipartite cells could normally form and conchospores would release. The low light with high density (HD) condition was set to explore high density effects under which most cells are in the hollow morph without bipartite cells formation and conchospore release. The high light with low density (HL) condition was set to explore high-light effects under which bipartite cell formation would be accelerated and conchospore release would be inhibited. Furthermore, we used brefeldin A (BFA), an inhibitor target of the BFA-inhibited guanine nucleotide exchange protein (BIG) gene, which is a key gene in vacuole formation-related vesicle traffic (Takagi and Uemura 2018), to further explore the mechanisms of hollow cell formation under high-density conditions. The results increase our understanding of sporangial filament development and serve as a foundation for the application of free-living sporangial filaments in production.
MATERIALS AND METHODS
Algal culture and group establishment
The original pure free-living sporangial filaments were preserved in the laboratories of the Institute of Oceanology, Chinese Academy of Sciences (Qingdao, China) at 23°C under 50 μmol photons m−2 s−1 irradiance (light : dark = 10 : 14) using PES medium (Provasoli 1958). The medium was replaced every 2 weeks, and the filaments were cut from the culture every 3 months.
To explore the mechanisms responsible for the effects of culture density and light intensity on sporangial filaments at the gene expression level, we established three groups, HD (cultured under low light intensity at high density), LD (cultured under low light intensity at low density) and HL (cultured under high-light intensity at low density), to conduct a transcriptome analysis. For the HD group, we placed 50 mg (fresh weight) of cut-off sporangial filaments into 50 mL culture medium (final culture density of approximately 13,000 fragments mL−1). For the LD group, we placed 50 mg (fresh weight) of cut-off sporangial filaments into 5 L of culture medium (final density of approximately 130 fragments mL−1). For the HL group, we placed 50 mg (fresh weight) of cut-off sporangial filaments into 5 L culture medium (final density of approximately 130 fragments mL−1). The light intensity for the HD and LD groups was adjusted to 10 μmol photons m−2 s−1, and the light intensity of the HL group was adjusted to 80 μmol photons m−2 s−1. The cutting of free-living sporangial filaments, and the relationship between fragments density and fresh weight, were carried out according to previously described method (He et al. 2021b).
All three groups were established in triplicate and cultured in an illuminating incubator (GXZ380B; Ningbo Jiangnan Instrument Factory, Ningbo, China) under a 10-h : 14-h light : dark photoperiod (18°C light : 21°C dark). The cultures were shaken three times every day. The materials were collected on the sixth day and frozen in liquid nitrogen. They were then stored at −80°C for RNA extraction.
RNA extraction, library construction, and sequencing
Total RNA was extracted using the RNAprep Pure Plant Kit (Tiangen, Beijing, China) according to the manufacturer’s instructions. Then, each RNA sample was analyzed by 1% agarose gel electrophoresis, and the quality and concentration of the RNA were evaluated using a NanoPhotometer spectrophotometer (Implen, Munich, Germany) and an RNA Nano 6000 Assay Kit on the Agilent Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA), respectively. A total of 1.5 μg RNA per sample was used as input material. Libraries were constructed using nine samples with grade A quality test results (highest grade for cDNA library construction). The original illumina sequencing was performed by Novogene Bioinformatics Technology Co., Ltd. (Beijing, China).
Bioinformatics analysis
After high-throughput sequencing, the raw reads were obtained. By removing low-quality sequences and adaptors, the clean reads were obtained and used for the subsequent analyses. The bioinformatics analysis is similar to the description in previous article (He et al. 2021a). Briefly, the clean reads were mapped onto the reference genome of Neopyropia yezoensis (Wang et al. 2020a) through HISAT2. 2.4 (Kim et al. 2015). Then using StringTie v1.3.1 (Pertea et al. 2016) to assemble the mapped reads of each sample. The RSEM (Li and Dewey 2011) was used to calculate the fragment per kilobase of transcript per million mapped reads (FPKM) value, which could quantify the expression abundance and variations of transcription regions. The differential expression analysis in two comparisons (HD vs. LD and HL vs. LD) was conducted by DESeq2 software (Love et al. 2014) with the differentially expressed genes (DEGs) screen thresholds: [log2 fold changes (FC)] ≥ 1 and corrected p-value (false discovery rate) < 0.05. Four databases (NCBI nonredundant protein sequences [NR], Universal Protein [Uniprot], Gene Ontology [GO], and Kyoto Encyclopedia of Genes and Genomes [KEGG]) were used for gene functional annotation. These analyses were conducted by another bioinformatic company (Genedenovo Biotechnology Co., Ltd., Guangzhou, China). KEGG pathway enrichment analysis of DEGs was performed using the OmicShare tools of Genedenovo Biotechnology Co., Ltd. (https://www.omicshare.com/tools).
Quantitative reverse transcription polymerase chain reaction validation of RNA-Seq
On the bases of the DEG analysis, seven DEGs were selected for validation by quantitative reverse transcription polymerase chain reaction (RT-qPCR). For the internal controls, 18S rRNA was selected. The details of RT-qPCR primers, which were designed using Primer 5.0 software, are listed in Supplementary Table S1. Each RT-qPCR was performed using an iQTM5 Multicolor Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) with Fast Start Essential DNA Green Master (Roche, Basel, Switzerland). The RT-qPCR mix for one 20 μL reaction contained the following components: 1 μL of cDNA template, 10 μL of 2× SYBR Green Master Mix, 1 μL of each primer (10 μmol μL−1), and 7 μL of RNase-free water. The program was set as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 10 s, 60°C for 15 s, and 72°C for 25 s, then 61 cycles of 65°C for 30 s. RT-qPCR was conducted for each gene, with three technical replicates. A melting curve analysis was performed to confirm product specificity. The relative gene expression values were calculated using the 2−ΔΔCt method (Livak and Schmittgen 2001).
BFA treatment
The BFA (APExBIO, Houston, TX, USA) treatment was conducted to determine whether the hollow morph of sporangial filaments was caused by vacuole fusion, as observed in previous research (He et al. 2021b), and regulated by the BIG gene detected in the transcriptome. BFA was dissolved in DMSO to a 10 mM final concentration. In the treatment group, sporangial filaments were cultured in PES medium containing a 10 μM final BFA concentration and a 0.1% volume ratio of DMSO, whereas the control groups were cultured in PES medium with or without a 0.1% volume ratio of DMSO. The initial culture density was approximately 1,300 fragments mL−1. Microscopic observations were conducted using an inverted microscope (Nikon, Tokyo, Japan), and fresh weights were measured after absorbing water with sterilized dust-free paper. Each treatment was performed with three biological replicates.
RESULTS AND DISCUSSION
Overview of transcriptome
After filtering low-quality data, an average 65.27 million (M) clean reads (with average = 91.59%) for nine samples were obtained (Table 1). After mapping to the reference genome (Wang et al. 2020a), the average mapped rate of each sample was 85.70% and the average exon ratio was 84.14%. The sample expression violin plot showed that the FPKMs of most genes were ranged in 0–100 and the FPKM median of all genes was around 10 (Supplementary Fig. S2) in nine samples. The correlation analysis indicated the good repeatability of samples within groups (Supplementary Fig. S3).
The DEGs were identified using two comparative analyses (HD vs. LD and HL vs. LD) with thresholds of q-value (p-adj) < 0.05 and |log2FC| > 1. In the HD vs. LD comparative group, there were 666 DEGs, with 279 DEGs upregulated in HD group and 327 DEGs downregulated in HD group, whereas in the HL vs. LD comparative group, there were 900 DEGs with 296 DEGs upregulated in HL group and 604 DEGs downregulated in HL group (Supplementary Fig. S4). The heatmap cluster of all the DEGs (Supplementary Fig. S5) showed that the gene expression pattern for HD samples was closer to that for LD samples compared with HL samples.
To confirm the reliability of transcriptome data, seven DEGs, including vegetative growth-related DEGs (ribulose phosphate kinase: py04528; carbonic anhydrases: py03023; phosphoenolpyruvate carboxylase: py09034; and nitrate reductase: py08787) and stress-related DEGs (universal stress proteins: py07868; betaine aldehyde dehydrogenase: py11381; and bromoperoxidase: py07165), were selected for verification. The expression trends of these DEGs were almost consistent with those in the transcriptome, indicating the high reliable of the transcriptome data (Fig. 1).

Verification of differentially expressed gene (DEG) levels. prkB, ribulose phosphate kinase; ald, betaine aldehyde de-hydrogenase; NR, nitrate reductase; cah, carbonic anhydrase; pepc, phosphoenolpyruvate carboxylase; LD, low light with low density group; HD, low light with high density group; HL, high light with low density group; RNA-Seq, RNA sequencing; RT-qPCR, quantitative reverse transcription polymerase chain reaction. Different lowercase letters indicate significant differences (t-test, p < 0.05).
Pathway enrichment of DEGs
Based on KEGG database, the pathway enrichment of DEGs in two comparative group were conducted (Fig. 2). For HL vs. LD comparative group, the changes mainly caused by light intensity, the 296 upregulated DEGs were enriched to many vegetative growth and substance synthesis-related pathways, such as “carbon metabolism”, “biosynthesis of secondary metabolites”, “biosynthesis of amino acids”, “glycine, serine and threonine metabolism”, “carbon fixation in photosynthetic organisms”, “photosynthesis - antenna proteins”, “nitrogen metabolism”, and “Pentose phosphate pathway” (Fig. 2A). These upregulated pathways further revealed the molecular basis of rapid growth of sporangial filaments under high light (80 μmol photons m−2 s−1) comparing with low light (10 μmol photons m−2 s−1) (He et al. 2021b). However, for the 604 downregulated DEGs of HL vs. LD comparative group, very few DEGs had been enriched (Fig. 2B), which meant the function of many of those DEGs could not been annotated in the KEGG database. For the downregulated DEGs, there were three DNA replication related pathways (“DNA replication”, “non-homologous end-joining”, “nucleotide excision repair”) appeared in top 10 enriched pathways (Fig. 2B), which the active DNA replication had also been observed in newly released conchospores from free-living filaments in our previous report (He et al. 2021a).

Pathway enrichment of differentially expressed genes (DEGs). (A) Pathway enrichment of upregulated DEGs in low light with high density group (HL) vs. low light with low density group (LD) comparative group. (B) Pathway enrichment of downregulated DEGs in HL vs. LD comparative group. (C) Pathway enrichment of upregulated DEGs in low light with high density group (HD) vs. LD comparative group. (D) Pathway enrichment of downregulated DEGs in HD vs. LD comparative group. The horizontal axis is rich factor. The vertical axis is the top 20 enriched pathways according to q-value.
For HD vs. LD comparative group, the changes mainly caused by culture density, the 279 upregulated DEGs were mainly enriched to vegetative growth-related pathways (“photosynthesis”, “nitrogen metabolism”, “photosynthesis - antenna proteins”, “starch and sucrose metabolism”, and “carbon metabolism”), stress response pathways (“cyanoamino acid metabolism”, “glyoxylate and dicarboxylate metabolism”, and “peroxisome”), and phospholipid metabolism (“sphingolipid metabolism”, “glycerolipid metabolism”, and “glycosphingolipid biosynthesis - globo and isoglobo series”) (Fig. 2C). For 327 downregulated DEGs of HD vs. LD comparative group, there were also a small amount of DEGs enriched, in which lipid metabolism pathways (“linoleic acid metabolism”, “biosynthesis of unsaturated fatty acids”, “fatty acid metabolism”, “fatty acid elongation”, “alpha-Linolenic acid metabolism”, and “fatty acid biosynthesis”) were the mainly enriched pathways (Fig. 2D).
Molecular basis of vegetative growth of sporangial filaments
According to KEGG enrichment (Fig. 2), the vegetative growth-related DEGs, which were mainly involved carbon dioxide concentration and fixation, photosynthesis, chlorophyll synthesis, and nitrogen absorption, had been collected. A heatmap of these DEGs (Fig. 3, details shown in Supplementary Table S2) showed that 12 carbon concentration- and fixation-related DEGs (carbonic anhydrases: py03023, py09926, and py04157; phosphoenolpyruvate carboxylase: py09034; aspartate aminotransferase: py09803; transketolase: py07592; ribulose phosphate kinase: py04528; phosphoglycerate kinase: py09381; fructose diphosphate aldolase: py04607; ribulose phosphate 3-epiisomerase: py05501; triose phosphate isomerase: py07449; and fructose-1,6-diphosphate esterase I: py10169), four light-harvesting protein DEGs (light-harvesting complex I chlorophyll a/b binding protein 1: py00271, py00479, and py06810; phycocyanin-associated rod linker protein: py03569), four photosynthesis DEGs (photosynthetic NDH subunit of subcomplex B3: py07502; Psb29: py02520; PGR5: py03620; and high light-inducible protein: py01375), three chlorophyll synthesis-related DEGs (protochlorophyllide reductase: py10059; glutamyl-tRNA reductase: py09283; and geranylgeranyl diphosphate a reductase: py03856), and two nitrogen absorption DEGs (nitrate transporter: py04172 and nitrate reductase: py08787) were upregulated in the HL group comparing with the LD group, indicating the molecular basis of rapid vegetative growth in sporangial filaments under high-light conditions, which had been observed previously (He et al. 2021b). In addition, some vegetative growth-related DEGs were upregulated in the HD group, including carbonic anhydrases (py00681 and py09926), fructose-1,6-bisphosphatase I (py10169), light-harvesting complex I chlorophyll a/b binding protein 1 (py00479 and py06810), gamma subunit of phycoerythrin (py02859 and py10352), photosystem II DEGs (psbM: py08479; psbO: py08802; and psbQ: py10045) and nitrate reductase (py08787), indicating that a high-culture density might promote the growth of sporangial filaments compared with a low-culture density.
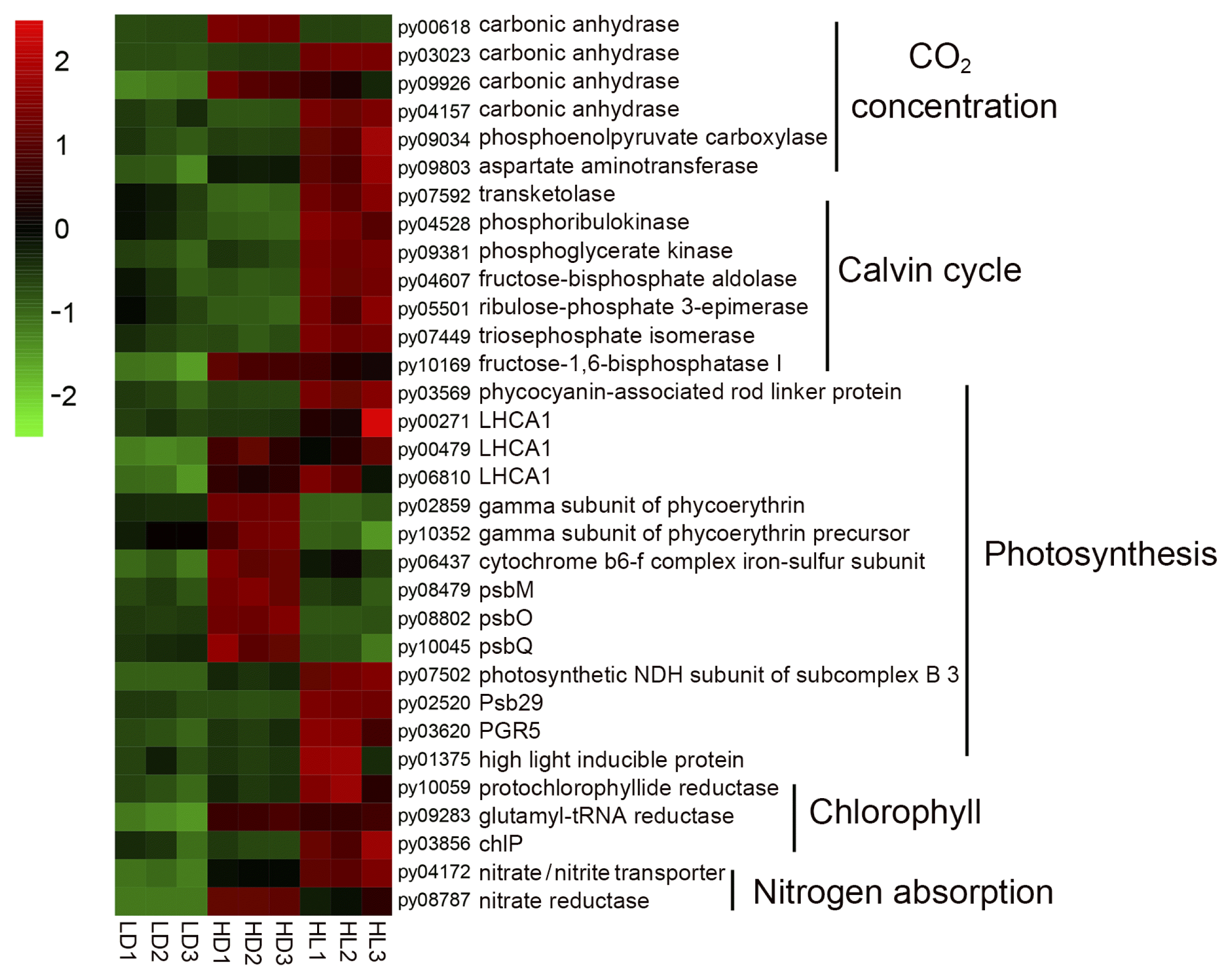
Heatmap of vegetative growth-related differentially expressed genes. Differential expression is shown in different colors. A negative number indicates the downregulation of genes, and a positive number indicates the upregulation of genes. LD, low light with low density group; HD, low light with high density group; HL, high light with low density group.
The difficulty in regulating conchospore release from free-living sporangial filaments was the main factor restricting the application of free-living sporangial filament-based seedling collection of Neopyropia / Neoporphyra. One important reason for the difficulty is the rapid vegetative growth of sporangial filaments in the free-living mode (He et al. 2021b). The matured bipartite cells (signs of sporangial filament maturation) could also return to the vegetative growth state. Hence, vegetative growth is the opposing process to conchospore release. For the current widely applied shell-boring sporangial filament seedling collection method, we speculated that a low vegetative growth rate might occur in shell-boring sporangial filaments because of the low-light condition in the shell. The low vegetative growth rate might be the main reason for the easy control of conchospore release from shell-boring sporangial filaments.
Molecular basis of the hollow morph in sporangial filaments under high-density conditions
The hollow morph of a sporangial filament in Neopyropia / Neoporphyra was first reported by Gui (Gui 1981), in which “cell starvation” was proposed as the main reason for the hollow morph. In our previous observations, the hollow morph was the main cell morphology of sporangial filaments under high-culture density conditions, in which bipartite cell formation is inhibited and nearly no conchospores are released, and the further ultrastructural observations showed that intracellular degradation and vacuolar expansion are responsible for the hollow morph (He et al. 2021b). In this transcriptome analysis, we found a key gene in vacuole formation-related vesicle traffic—BFA-inhibited guanine nucleotide exchange protein (BIG, py05721) (Kitakura et al. 2017, Takagi and Uemura 2018), and it was expressed highest in the HD group (Fig. 4). Moreover, we used BFA, an inhibitor of vesicle traffic and the target of BIG, to treat the sporangial filaments. The morphological changes (Fig. 5) revealed that the BFA-treated group produced an abnormal solid morph compared with the normal hollow morph in the control group. When BFA was removed to test recovery, new hollow tip cells occurred by the second day and the normal hollow morph occurred by the ninth day. In contrast, the sporangial filaments still being treated with BFA (BFA-26 day) produced the abnormal solid morph. Fresh weight changes (Supplementary Fig. S6) showed that the BFA treatment also significantly inhibited the vegetative growth of sporangial filaments both under low- and high-light conditions. These results strongly support the hollow morph being directly influenced by vacuole formation-related vesicle traffic.
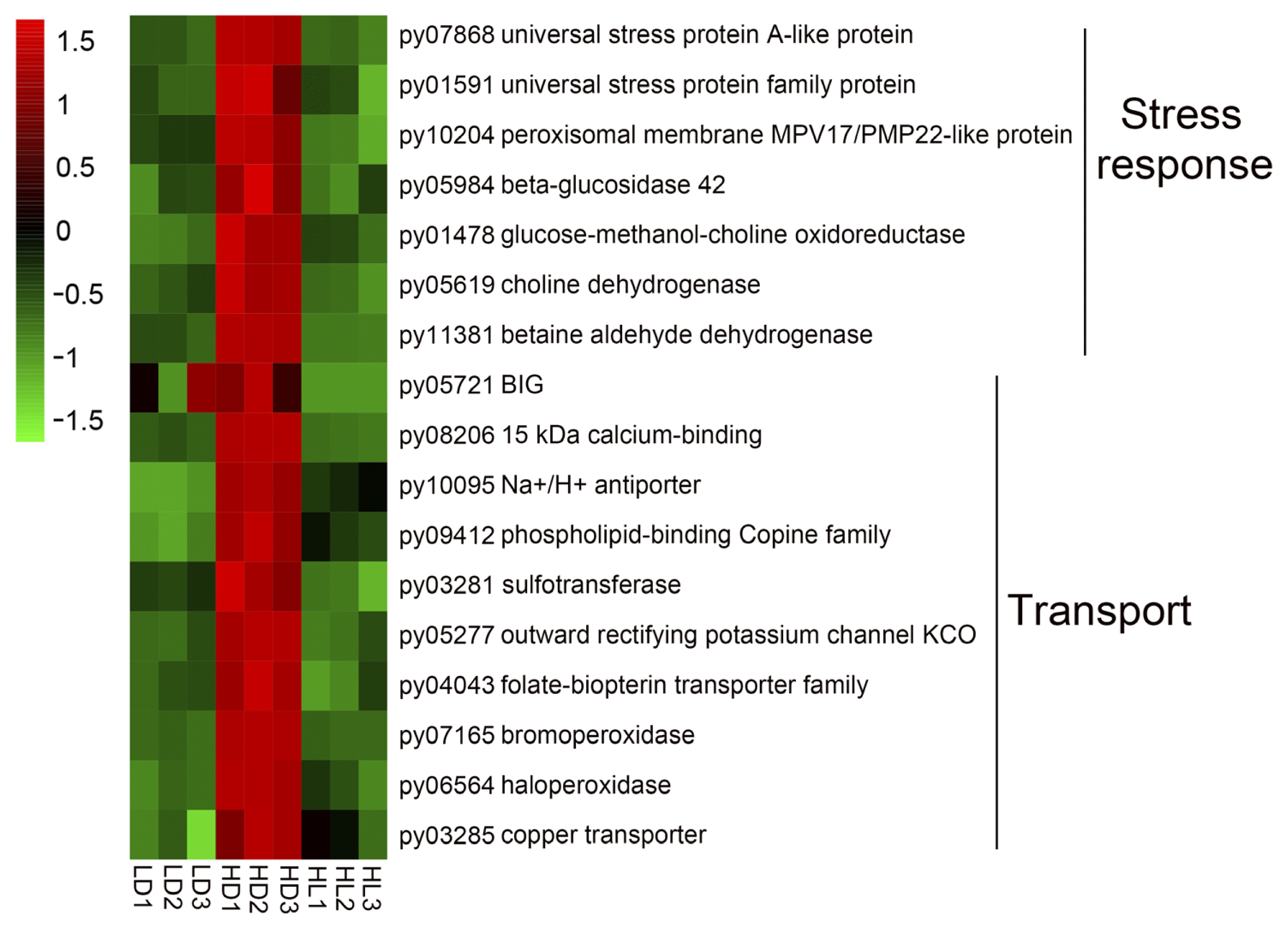
Heatmap of stress- and ion transport-related differentially expressed genes. Differential expression is shown in different colors. A negative number indicates the downregulation of genes, and a positive number indicates the upregulation of genes. BIG, brefeldin A-inhibited guanine nucleotide exchange protein; LD, low light with low density group; HD, low light with high density group; HL, high light with low density group.
Morphology of sporangial filaments under brefeldin A (BFA)-treatment conditions. The treatment was a 10 μM final BFA concentration with a 0.1% volume ratio of DMSO. The control group was treated with a 0.1% volume ratio of DMSO only. Recovery started on the 17th day after BFA treatment. Thus, the 26th day of the BFA treatment was the control of 9th day of recovery. Arrows indicate newly formed normal hollow tip cells. Scale bars represent: 20 μm.
In addition, we found that many of the stress response- and transport-related DEGs were expressed highest in the HD group (Fig. 4, details shown in Supplementary Table S2), mainly caused by high-culture density. The stress response DEGs contained salt and osmotic stress-related genes, such as universal stress proteins (py07868 and py01591) (Udawat et al. 2016), peroxisomal membrane MPV17-like protein (py10204) (Wi et al. 2020), betaine aldehyde dehydrogenase (py11381) (Ishitani et al. 1995), and environmental stress-related cyanide metabolism DEGs (beta-glucosidase: py05984; glucose-methanol-choline oxidoreductase: py01478; and choline dehydrogenase: py05619) (Xu et al. 2012). The transport DEGs were involved in transport of calcium (15 kDa calcium-binding: py08206), Na+/H+ (Na+/H+ antiporter: py10095), phospholipid (phospholipid-binding copine family: py09412), sulfo (sulfotransferase: py03281), K+ (outward rectifying potassium channel KCO: py05277), folate-biopterin (folate-biopterin transporter family: py04043), bromide (bromoperoxidase: py07165), halogen (haloperoxidase: py06564), and Cu2+ (copper transporter: py03285) (Fig. 4).
The highest DEGs in the HD group indicated that high-density culture conditions caused stress pressure, mainly related to salt and osmotic stresses, and the exchange between sporangial filament cells and the extracellular medium may be much more active compared with under low-density culture conditions. Thus, the developed vacuoles may be needed to regulate salt exchange and storage. Finally, the hollow morph of free-living sporangial filaments formed under high-culture density conditions. Based on these molecular results, we proposed that the difficulties associated with maturation and conchospore release caused by the high-density culturing of free-living sporangial filaments might be overcome by alleviating salt and osmotic stresses.
Potential conchospore release-related genes
In the life cycle of N. yezoensis, spore releasing, including conchospore, archeospore and zygotospore releasing, were important phenomena (Supplementary Fig. S1). Many studies have reported the effects of different environmental factors and treatments on conchospore (Chen 1980, He and Yarish 2006, Li et al. 2011) and archeospore (Mizuta et al. 2003, Blouin et al. 2007, Takahashi and Mikami 2017, Chen et al. 2019) release. However, little is known about the molecular bases of spore releasing. For the three conditions, conchospores could be released normally in LD group but the release was inhibited in both the HD and HL groups (He et al. 2021b). Thus, we determined the highest DEGs which were in the LD group to explore the potential molecular basis of conchospore release. Those DEGs are shown in Fig. 6 (details shown in Supplementary Table S2), including zinc-finger proteins (py02218, py05773, py01069 and MSTRG.6889), cell wall component genes (xylosyltransferase: py05402 and py08274; glycosyltransferase: py04239 and py07945; lpxtg-motif cell wall anchor domain protein: py07099), ASPO1527 (py01333, py08113, py08112, py10361, and py11032), cell cycle genes (cyclin-U2-2: py00664 and pachytene checkpoint protein: py00925) and microtubule-related genes (tubulin-folding cofactor: py10326 and severin: py09077). In plants, the zinc-finger protein family has many effects on plant growth and development (Li et al. 2013). ASPO1527 was identified by Kitade et al. (2008), and it might be associated with archeospore formation and release. Here, we speculated that it may also be associated with conchospore release. The DEGs involved in cell wall component, cell cycle and microtubule pathways may be related to conchospore formation and movement in the tubes of sporangial filaments.

Heatmap of differentially expressed genes potentially related to conchospore release. Differential expression is shown in different colors. A negative number indicates the downregulation of genes, and a positive number indicates the upregulation of genes. LD, low light with low density group; HD, low light with high density group; HL, high light with low density group.
CONCLUSION
This research explored the molecular bases of vegetative growth, hollow morph, and conchospore release of free-living sporangial filaments controlled by light levels and culture density. High light promoted the expression of DEGs related to carbon dioxide concentration and fixation, photosynthesis, chlorophyll synthesis and nitrogen absorption, and hence, promoted the growth of sporangial filaments. High density caused the upregulation of a series stress response- and transport-related DEGs, especially the upregulation of a key gene involving in vacuole formation-related vesicle traffic, and hence, caused the hollow morph of sporangial filaments. The DEGs annotated as zinc-finger proteins, cell wall component, cell cycle, microtubule, and ASPO1527, might be associated with conchospore formation and release. These results increase our understanding of the molecular mechanisms of sporangial filament development and may aid in the artificial collection of conchospores from N. yezoensis.
ACKNOWLEDGEMENTS
This work was supported by the National Natural Science Foundation of China (32202907, 42376091, 42276146), the Major Scientific and Technological Innovation Project of Shandong Provincial Key Research and Development Program (2022LZGC004), the Research Fund for the Taishan Scholar Project of Shandong Province (tspd20210316), and China Agriculture Research System of MOF and MARA (CARS-50).
Abbreviations
BFA
brefeldin A
BIG
brefeldin A-inhibited guanine nucleotide exchange protein
DEGs
differentially expressed genes
FC
fold changes
FPKM
fragment per kilobase of transcript per million mapped reads
HD
low light with high density group
HL
high light with low density group
LD
low light with low density group
RT-qPCR
quantitative reverse transcription polymerase chain reaction
Notes
The authors declare that they have no potential conflicts of interest.
SUPPLEMENTARY MATERIALS
The life cycle of Neopyropia yezoensis (https://www.e-algae.org).
Sample expression violin plot (https://www.e-algae.org).
Heat map of sample correlation (https://www.e-algae.org).
Numbers of differentially expressed genes in two comparisons (https://www.e-algae.org).
Clustering of all the differentially expressed genes (https://www.e-algae.org).
Effects of brefeldin A (BFA) treatment on fresh weights of sporangial filaments (https://www.e-algae.org).
Primers used in RT-qPCR (https://www.e-algae.org).
Details of all DEGs involved in this article (https://www.e-algae.org).
