INTRODUCTION
Coccolithophores are major phytoplankton species among the haptophyta lineage that produce large blooms in oceans and contribute to global ocean calcification (Taylor et al. 2017). Emiliania huxleyi is the most abundant coccolithophore species found in oceans (Westbroek et al. 1993). It is known to produce delicate calcium carbonate scales called coccoliths (Westbroek et al. 1989). Coccoliths are produced in an intracellular compartment called a coccolith vesicle (CV), which is derived from the Golgi apparatus. Concentrating calcium ions and carbonates in the CV is a key process for establishing the geometric shapes of the calcium carbonate structure (Westbroek et al. 1993). However, molecular mechanisms underlying the elaborate calcification process remain unclear.
von Dassow et al. (2009) compared the transcriptomes of haploid non-calcifying cells and diploid calcifying cells and identified several genes in the diploid cells that are potentially involved in calcification. However, the differential gene expression patterns observed could also be related to differences between haploid and diploid stages (von Dassow et al. 2009). Mackinder et al. (2011) analyzed the transcript levels of eight putative genes related to calcification using quantitative reverse transcription polymerase chain reaction (qRT-PCR). In particular, they compared gene expression between two isogenic pairs of E. huxleyi (RCC 1216 vs. RCC 1217 and CCMP 1516 vs. CCMP 1516NC) and found that genes encoding a putative HCO3− transporter, Ca2+/H+ exchanger, and vacuolar H+-ATPase were upregulated in the calcifying strain compared with the non-calcifying strain (Mackinder et al. 2011). Comparative studies have identified several putative calcification-related genes and are a foundation for further research on the calcification of E. huxleyi. However, the underlying molecular basis of calcification is still unclear.
Coccolith production by E. huxleyi can vary by the composition of the culture medium. Previous studies have demonstrated that growing E. huxleyi at different calcium concentrations altered coccolith formation by calcifying cells (Herfort et al. 2002, 2004, Trimborn et al. 2007, Leonardos et al. 2009, Mackinder et al. 2011). E. huxleyi strains grown in a calcium-free culture medium did not form coccoliths. Conversely, cells cultured at an ambient concentration of calcium (10 mM) in the culture medium were enclosed in coccoliths. In this study, to gain further insight into coccolith production, we compared the transcriptomes of a calcifying (CCMP 371) and a non-calcifying (CCMP 2090) strain of E. huxleyi and identified candidate calcification-associated genes. Then, we examined the expression of a subset of those putative calcification-associated genes in CCMP 371 grown at different calcium concentrations.
MATERIALS AND METHODS
Algal strains and culture conditions
Calcifying (CCMP 371) and non-calcifying (CCMP 2090) algal strains were purchased from the Provasoli-Guillard National Center for Marine Algae and Microbiota. Both strains were cultured in sterile, filtered artificial seawater (ASW) (Soto et al. 2006) enriched with nitrates (final concentration 880 μM), phosphates (final concentration 36 μM), trace metals, and vitamins at f/2 concentration (Guillard 1975), with selenium (final concentration 0.01 μM) (Araie et al. 2011). Calcium concentrations used in the experiments for CCMP 371 were 0, 0.1, and 10 mM. Cells were agitated twice a day and maintained at 20°C photoautotrophically under irradiance of 40 μmol−1 m−2 (12: 12 h light: dark). Growth was monitored by counting cells using a hemocytometer (Marienfeld, Bad Mergentheim, Germany). All experiments were performed with cells in the exponential stage of growth.
Sample preparation
Cultures maintained in the exponential growth phase were inoculated in fresh media at a cell concentration of 1 × 106 cells mL−1. The cell concentration was monitored by counting cells using a hemocytometer. For all experiments, cells were harvested at day 4 of the growth phase. On day 4, density of cells cultured in different calcium concentrations 0, 0.1, and 10 mM were 6.81 × 106, 7.85 × 106, and 7.88 × 106 cells mL−1, respectively.
Transcriptomic analysis
Cells for transcriptome analysis were maintained at 20°C photoautotrophically under irradiance of 40 μmol−1 m−2 (12: 12 h light: dark) agitated twice a day by hand shaking. Cells in the exponential phase of the growth were used for RNA extraction. Fifty mililiteres of each cell cultures (CCMP 371 and CCMP 2090) were harvested by centrifugation (2,170 ×g, 10 min). After discarding the supernatant, pellet was immediately frozen using liquid nitrogen and kept on -80°C for RNA extraction. The total RNA from CCMP 371 and CCMP 2090 were extracted using the Hybrid-R kit (GeneAll, Seoul, Korea) following the manufacturer’s instructions. Contaminating DNA was eliminated using RNase-Free DNase Set (Qiagen, Hilden, Germany). Extracted total RNA was sent to Macrogen (Seoul, Korea) for library construction, sequencing, and assembly using the HiSeq 2000 system (Illumina, San Diego, CA, USA). Genes that were significantly differentially expressed between calcifying and non-calcifying strains were analyzed by Macrogen. The sequence information for CCMP 371 (accession SRR5894809) and CCMP 2090 (accession SRR5894808) was deposited in the National Center for Biotechnology Information database.
RNA preparation, cDNA synthesis, and qRT-PCR
The total RNA was extracted from cell cultures using the Hybrid-R kit (GeneAll). cDNA was synthesized using the Superscript III kit (Invitrogen, Carlsbad, CA, USA) primed with oligo dT primers by following the manufacturer’s instructions. The cDNA was used as a template for qRT-PCR that used SYBR green chemistry for amplicon detection. An SYBR premix (Takara, Tokyo, Japan) and the Thermal Cycler Dice Real Time System TP8200 (Takara) were used for cDNA amplification. Actin was used as the reference gene in qRT-PCR (Mackinder et al. 2011). Sequences of primers for the target and reference genes are provided in Table 1. Melt curves were checked to confirm there was no non-specific priming in samples.
RESULTS
Relative gene expression of putative calcification genes
Many studies have attempted to clarify the calcification process at the molecular level by comparing gene expression patterns between coccolith- and non-coccolith-producing E. huxleyi strains using expressed sequence tags, suppressive subtractive hybridization, serial analysis of gene expression, and cDNA microarrays (Wahlund et al. 2004a, 2004b, Nguyen et al. 2005, Dyhrman et al. 2006, Quinn et al. 2006, von Dassow et al. 2009). In this study, we used high-throughput RNA sequencing (HiSeq2000; Illumina) to elucidate genes associated with biomineralization.
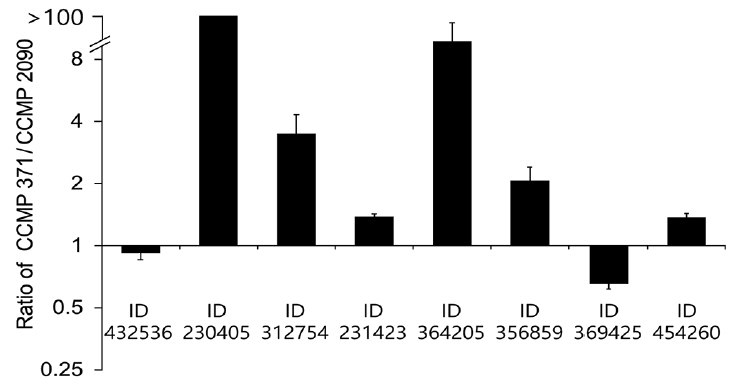
The transcriptomes of the calcifying (CCMP 371) and non-calcifying (CCMP 2090) strains were compared to identify potential genes associated with calcification (Supplementary Fig. S1). The genes were selected from the highly expressed transcripts in CCMP 371 compared to CCMP 2090. In our transcriptome data 35141 contigs showed highly differentially expressed in CCMP 371 over 2-fold. By using BLAST search (BLASTX against a non-redundant protein database), among these contigs 5125 did not show any match with other published databases (no hit). Twenty-four thousand seven hundred twenty-nine contigs were hypothetical proteins and 5,287 contigs were showing a homology with the annotated genes including E. huxleyi and other species. Four hypothetical genes most highly differentially expressed in CCMP 371 compared to CCMP 2090 were selected for the study. For the other four differentially expressed putative genes, we selected the genes which were speculated to be related to cytoskeleton, translation, bicarbonate transporter and carbonic anhydrase that were not investigated from other published reports. We selected eight candidate genes that were upregulated in CCMP 371 compared with that in CCMP 2090 (Supplementary Fig. S2) for further validation via qRT-PCR (Fig. 1). Genes known to be related to biomineralization reported by Mackinder et al. did not showed an increased expression in CCMP 371 compared to CCMP 2090 from our transcriptome data (Supplementary Table S1). Four putative genes were relatively highly expressed in CCMP 371 compared with those in CCMP 2090. Other candidate genes showed relatively low expression or were slightly differentially expressed between the calcifying and non-calcifying strains.
Since the strains are not isogenic, we investigated the conditions that could modulate the calcifying ability of CCMP 371. To eliminate effects related to isogenicity, we cultured CCMP 371 at different calcium concentrations that could potentially regulate coccolith production (Herfort et al. 2004, Trimborn et al. 2007, Leonardos et al. 2009).
Culture of CCMP 371 at different Ca2+ concentrations
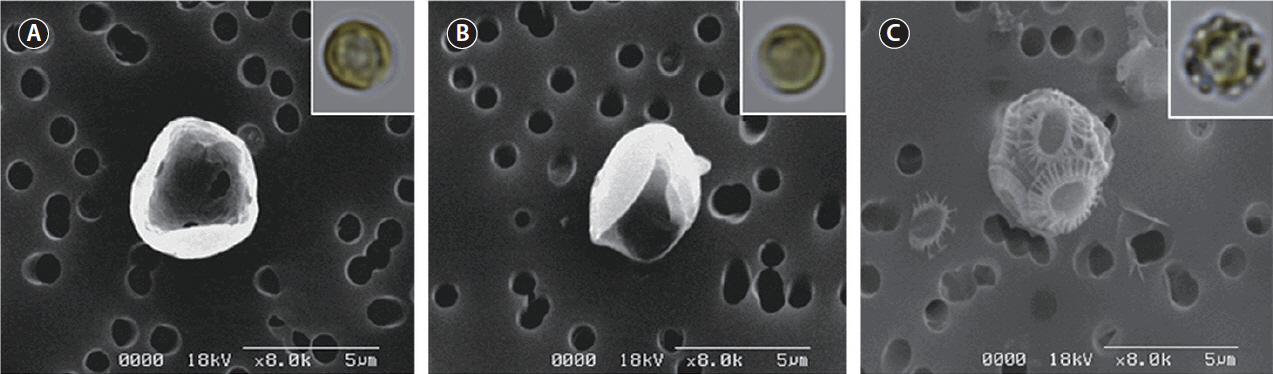
To modulate the calcifying state of CCMP 371, we first adapted the cells to specific calcium concentrations. Cells were subcultured at least five times at the indicated calcium concentration before the experiment. Cells cultured in ASW with an ambient concentration of calcium (10 mM) displayed intact coccoliths when examined under SEM (Fig. 2C). Conversely, cells cultured in the absence or at a low concentration (0.1 mM) of calcium did not display coccoliths (Fig. 2A & B). These results are consistent with previous reports that evaluated coccolith production at different calcium concentrations using other E. huxleyi strains (Herfort et al. 2004, Trimborn et al. 2007, Leonardos et al. 2009).
Growth of CCMP 371 at different Ca2+ concentrations
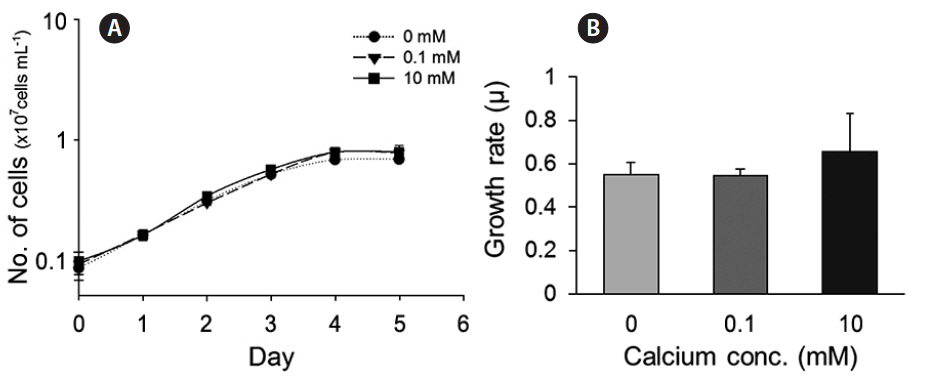
We also evaluated the growth of cells in culture medium with different concentrations of calcium (0, 0.1, and 10 mM). Cells cultured under calcium-limiting conditions (0 and 0.1 mM) did not show calcium-dependent growth, i.e., growth was not limited by very low calcium (Fig. 3A & B).
Relative expression of putative calcification genes in CCMP 371 cells grown at different Ca2+ concentrations
To check whether the expression of putative genes is correlated with calcium concentrations, we have verified the gene expression level in CCMP 371 by modulating the coccolith formation. Cells were harvested at the exponential growth phase, and the total RNA was subsequently isolated for qRT-PCR. Candidate genes encoding hypothetical protein (Protein ID 230405) (Fig. 4A), translation-related protein eukaryotic initiation factor-4A (eIF4A) (Fig. 4B), and putative ATP-binding cassette (ABC) transporter (Fig. 4D) showed a calcium-dependent gene expression pattern. The translation-related protein eIF4A (Protein ID 312754) level was approximately 60% upregulated at an ambient calcium concentration than under calcium-limiting concentrations (Fig. 4B). In contrast, the cytoskeleton-related protein profilin (Protein ID 364205) (Fig. 4C) showed a low level of expression (four-fold decrease in expression) at an ambient calcium concentration than under calcium-limiting conditions. Putative carbonic anhydrase (CA) (Protein ID 356859) (Fig. 4E) and RUBISCO large subunit (rbcL) (Fig. 4F) did not show significant up or downregulation at different calcium concentrations.
DISCUSSION
CCMP 371 (strain synonym 12-1) was first isolated from the Sargasso Sea in the North Atlantic Ocean. CCMP 2090 is an axenic isolate of CCMP 1516 (isolated from the South Pacific). So far, CCMP 2090 has not appeared to calcify under any conditions (Soto et al. 2006), indicating that the two strains CCMP 371 and CCMP 2090 are not isogenic. However, E. huxleyi, which is globally distributed, has a pan genome (Read et al. 2013). Furthermore, this is the first time that the relative expression of putative calcification-associated genes in CCMP 371 and CCMP 2090 has been compared.
The morphological and physiological results show that it is appropriate to examine putative genes related to biomineralization under different calcium concentrations. These results are consistent with the experiments performed using the E. huxleyi strains PCC. B11, B92/11, and PML B92/11A (Herfort et al. 2004, Trimborn et al. 2007, Leonardos et al. 2009). Herfort et al. (2004) examined PCC. B11 with medium containing up to 20 mM calcium. Calcification rates increased whilst those of photosynthesis remained constant over the same range of calcium concentrations. Trimborn et al. (2007) has grown strain B92/11 which did not calcified under 0.1 mM calcium concentration and the growth rate did not show a significant decrease under irradiance of 30 μmol−1 m−2. Leonardos et al. (2009) examined the mechanistic dependence of photosynthesis on calcification in strain PML B92/11A. Previous studies show that calcifying E. huxleyi strains cultured under 0 and 0.1 mM calcium concentrations resulted in no production of coccoliths which corresponds to our results. Light-dependent O2 evolution measurements tested by Leonardos et al. (2009) showed lower calcium concentration had no evidence for lower photosynthetic performance. In contrast, CCMP 2090 did not calcify at ambient (10 mM) and low (0.1 mM) calcium concentrations (Supplementary Fig. S3).
Transcriptome levels of the candidate genes in CCMP 371 differed among cells exposed to 0, 0.1, and 10 mM calcium concentrations. Transcriptome levels of a hypothetical protein (Protein ID 230405) increased with the calcium concentration (Fig. 4A). This suggested that the protein encoded by this gene is involved in calcification or calcium homeostasis.
Eukaryotes can regulate gene expression by controlling translation rates. Translation initiation is catalyzed by eIF4A1, eIF4A2, and eIF4A3. These factors are necessary for the binding of mRNA to 40S ribosomal subunits (Gingras et al. 1999). In our study, the translation-related protein eIF4A was also highly expressed at an ambient (10 mM) calcium concentration than under calcium-limiting conditions, indicating that eIF4A shows calcium-inducible gene expression. The eIF4A level was upregulated by approximately 60% in the calcifying state compared with that in the non-calcifying state (Fig. 4B). Although the expression pattern showed a calcification-associated response at different calcium concentrations, further studies are required to validate the observed expression pattern of this gene under calcification conditions.
ABC transporters are one of the largest transport system superfamily in extant phyla. They are integral membrane proteins that translocate solutes across the cell membrane by employing the energy of ATP hydrolysis (Higgins 1992, Jones and George 2004). Bicarbonate uptake driven by the ABC transporter was identified in Synechococcus sp. strain PCC 7942 (Omata et al. 1999). In addition, bicarbonate transport is important for coccolith formation, and calcium uptake has been stimulated at a high concentration of bicarbonate (Fukuda et al. 2014). Although the function of the putative ABC transporter gene needs to be examined, its role can be speculated as a calcium-inducible gene related to calcification. In this study, the ABC transporter (Protein ID 231423) showed a calcium-inducible expression pattern and an approximately 37% lower transcriptome level under calcium-limiting conditions (Fig. 4D) compared with the ambient calcium (10 mM) concentration.
The cytoskeleton-related protein profilin (Protein ID 364205) showed an opposite expression pattern. Profilins are small actin-binding proteins involved in dynamic turnover of the actin cytoskeleton that is essential for all organisms. Profilins catalyze the exchange of ADP for ATP on actin monomers during actin polymerization. Profilins have also been reported to be involved in organelle location and vesicle trafficking (Revenu et al. 2004, Witke 2004, Sun et al. 2013). A previous study reported that treatment of algae with inhibitors of actin and microtubules resulted in malformed coccoliths (Langer et al. 2010). However, to uncover the particular roles of cytoskeletal structures in regulating calcification, further research is required. Cytoskeletal regulation must play a vital role in coccolith production because the cytoskeleton interacts with the membrane-trafficking system. From targeted trans-Golgi trafficking to association with the CV, the endomembrane system is tightly regulated to supply the organic and inorganic substrates for calcification. The cytoskeletal machinery controls the vesicle and cellular movements associated with the formation and secretion of coccoliths (Taylor et al. 2017). We observed a four-fold decrease in the gene expression of profilin under calcifying conditions compared with that under non-calcifying conditions (Fig. 4C). This lower expression of profilins at an ambient (10 mM) calcium concentration requires further investigation.
The putative CA (Protein ID 356859) (Fig. 4E) did not show a significant change at different calcium concentrations. CAs are ubiquitous metalloenzymes that catalyze the reversible hydration of CO2 and HCO3− in many organisms (Badger and Price 1994). Soto et al. (2006) characterized the function of γ-EhCA2 in vitro and speculated its location in the CV; this study was the first to characterize the CA isoform in E. huxleyi. RCC 1216 cells grown at an elevated partial pressure of carbon dioxide resulted in significantly reduced (3.8-fold) γ-EhCA2 transcripts, but calcification rates did not change significantly between the treatments (Richier et al. 2011). On the other hand, γ-EhCA2 showed 1.71-fold-increased gene expression in the calcifying conditions of CCMP 1516 (Mackinder et al. 2011). Still, the molecular features of multiple CA isozymes are scarce in microalgae and notably in E. huxleyi. Our results did not show calcium-inducible expression of putative CA, and characterizing the molecular function of the putative CA requires further analysis.
To summarize, there were differences in coccolith formation by CCMP 371, the calcifying strain of E. huxleyi, at different calcium concentrations, but growth was not affected by the calcium concentration. These results suggest that determining the expression of the three putative genes at the three different calcium concentrations described before is a reasonable approach for validating these candidate genes.
Several other factors affect the calcification of E. huxleyi, including phosphate deficiency and cold stress conditions (Kayano and Shiraiwa 2009, Satoh et al. 2009). According to Satoh et al. (2009), phosphate deprivation is the primary factor that triggers coccolith formation, whereas the secondary factor is low temperature. Therefore, validation of candidate genes under phosphate deficiency and cold stress conditions may provide additional information.