ABSTRACTDiatoms, a type of microalgae distributed worldwide, have been identified as potential sources of biomass, lipids, and high-value compounds. While marine diatoms have been extensively studied, the potential of freshwater diatoms still needs to be explored. In this study, a novel strain of freshwater diatom was isolated from the Jungnangcheon stream located in Seoul, Republic of Korea (37°33′08.0″ N, 127°02′40.0″ E). This newly isolated strain was classified through phylogenetic analysis, and its morphology was investigated using light and electron microscopy; it was named Nitzschia palea HY1. N. palea HY1 grown in freshwater media (FDM) produced higher biomass (0.68 g L−1) and fucoxanthin production (9.19 mg L−1) than in conventional diatom media. Furthermore, increasing the bicarbonate concentration from 2 to 10 mM enhanced the maximum biomass and fucoxanthin production in FDM by 2.7 fold and 1.5 fold, respectively. Remarkably, the introduction of aeration to the modified FDM (MFDM) led to a substantial increase in the maximum biomass and fucoxanthin production of N. palea HY1, exhibiting 3.8-fold and 4.1-fold enhancement, respectively, compared to FDM alone. These findings suggest that optimizing the cultivation of N. palea HY1 using MFDM could provide an alternative to marine sources for fucoxanthin production.
INTRODUCTIONMicroalgae can capture CO2 through photosynthesis, producing biomass and valuable resources (Jeong et al. 2023). Diatoms are one of the most abundant photosynthetic microalgae in fresh and marine environments and responsible for global primary production (Smetacek 1999, Finlay et al. 2002). Diatoms are also the main drivers of the biogeochemical cycles of silicon and other major elements in aquatic environments (Nelson et al. 1995, Tréguer et al. 2018). Moreover, they can efficiently produce valuable chemicals, such as omega-3 fatty acids, pigments, and chrysolaminarin (Yang et al. 2020). Fucoxanthin is a brown pigment that belongs to the xanthophyll category of carotenoids and is produced by brown algae and diatoms. Owing to its distinct chemical properties, this pigment is recognized for several biological functions with significant medicinal applications, such as anti-inflammatory, anti-obesity, anti-diabetic, anti-oxidant, anti-cancer, anti-microbial, anti-osteoporotic, and anti-hypertensive properties (Xiao et al. 2020, Seth et al. 2021).
Currently, fucoxanthin is primarily obtained from discarded parts of brown macroalgae, including Laminaria japonica, Eisenia bicyclis, Undaria pinnatifida, and Hijikia fusiformis. However, these macroalgae are primarily harvested for human consumption in Asia and contain low levels of fucoxanthin (Kanazawa et al. 2008). In contrast, microalgae are a promising source of fucoxanthin for commercial production as they have a much higher concentration of fucoxanthin than macroalgae, and the biotechnology for microalgae production is continuously advancing (Khaw et al. 2022). Despite the abundance and variety of fucoxanthin-producing microalgae, only a few have been investigated for commercial production of fucoxanthin (Kim et al. 2012, Xia et al. 2013, Khaw et al. 2022).
To date, there have been many challenges to efficiently producing biomass and fucoxanthin from diatoms by optimizing culture conditions, genetic manipulation, and isolating new strains with the desired properties (Seth et al. 2021). Although marine diatoms have been extensively researched for their potential as feedstock for various bioactive compounds, studies on optimizing the growth and biomass production of freshwater diatoms are limited (Griffiths and Harrison 2009).
Freshwater diatoms play an important ecological role as bioindicators of water quality because they are highly diverse and sensitive to environmental conditions (Vilmi et al. 2015, Chonova et al. 2019). Although freshwater diatoms share some features with marine diatoms, their low-salinity habitats have unique physiological and metabolic properties that require further investigation (Kilham and Hecky 1988, Litchman et al. 2009, Maberly et al. 2021). Therefore, it is necessary to optimize the growth and physiological properties of freshwater diatoms.
Most freshwater diatoms are cosmopolitan, and Nitzschia palea is a widely distributed benthic freshwater diatom (Trobajo et al. 2009, Tiam et al. 2018). It was previously reported that 26 strains from Belgium, Brazil, Egypt, India, Japan, Paraguay, Spain, Sri Lanka, and the United Kingdom were classified and named N. palea. However, they are an assortment of morphologically indistinguishable cryptic species (Trobajo et al. 2009). Although the worldwide presence of N. palea is known, few studies have investigated the physiology or metabolism of N. palea (Gérin et al. 2020), and only one case of its complete organelle genome sequence has been reported (Crowell et al. 2019).
Therefore, in this study, a Korean domestic microalga, N. palea was isolated from the Jungnangcheon stream in Seoul, Republic of Korea, and named strain HY1. This new freshwater strain was characterized morphologically and phylogenetically to determine its taxonomic identity. The growth conditions for N. palea HY1 were optimized to maximize fucoxanthin production.
MATERIALS AND METHODSIsolation and maintenance of Nitzschia palea
Nitzschia palea HY1 was isolated from the Jungnangcheon stream, Seoul, Republic of Korea (37°33′08.0″ N, 127°02′40.0″ E). Samples were collected from stone surfaces using a brush. Single living epilithic diatom cells were isolated using the capillary method (Andersen and Kawachi 2005) with an inverted microscope (CKX41; Olympus, Tokyo, Japan) and a glass Pasteur pipette (Hilgenberg GmbH, Malsfeld, Germany). Cells were isolated and cultured in 96-well cell plates, and each well contained 160 μL diatom medium (DM) (Beakes et al. 1988). Healthy cells that grew were transferred to 24-well cell plates with 1 mL of DM. After 7–10 days, the cells were multiplied and transferred to 50 cm3 culture flasks containing 20 mL of DM. Stock cultures were maintained at 20°C, under white fluorescent lamps at 100 μmol m−2 s−1 cool and a 12 : 12 h light : dark cycle.
In this study, N. palea HY1 was cultivated in freshwater diatom medium (FDM) (Gérin et al. 2020). Cells were grown at 20°C in 250 mL cell culture flasks with filter caps, under cool white fluorescent light at 100 μmol photons m−2 s−1 and a 12 : 12 h light : dark cycle, and the agitation rate was fixed at 130 rpm. For aeration of the culture media, filter-sterilized (0.2 μm syringe filters; Advantec, Tokyo, Japan) atmospheric air was bubbled through the culture medium (80 mL min−1).
Morphology of cell and its bio-silica structureBright-field and fluorescence images were obtained using an optical microscope (ECLIPSE Ni-u; Nikon, Tokyo, Japan) equipped with a metal halide illuminator (Photofluor LM-75; 89 North, Williston, VT, USA). Both images were captured using DS-Fi3 (Nikon) and merged using NIS-Elements version 4.60 build 1170 (Nikon). The specimens were then mounted on glass slides and observed.
Brown diatom pellets were isolated from the suspended cultures at 13,000 rpm to prepare and extract the bio-silica structure from the diatoms for electron microscopy. EDTA-SDS was used to remove organic constituents of the diatom frustules. Distilled water was used to wash diatom frustules. Intracellular components were extracted with methanol to minimize the impact of extraction on the chemistry and morphology of frustules. Each step was repeated several times to obtain clean, intact diatom frustules. Scanning electron microscopy (SEM) samples were deposited onto aluminum foil or silicon wafers and dried overnight. Each sample was examined using Apreo S Hivac (Thermo Fisher Scientific, Waltham, MA, USA) at Hanyang University.
DNA extraction and PCR amplificationFor DNA extraction, we placed 5 mL of clonally cultured samples in the mid-logarithmic growth phase into conical tubes and harvested the cells by centrifugation at 3,500 ×g for 10 min. Genomic DNA was isolated from the stored cells using a DNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA). PCR reactions were performed in 20 μL reaction mixtures, and the primers used in the PCR amplification of the small subunit ribosomal RNA (SSU rRNA) and rbcL genes are shown in Table 1.
The SSU rRNA reaction mixture contained 11.9 μL of sterile distilled water, 2 μL of dNTP (TaKaRa, Tokyo, Japan), 1 μL of each primer, 0.1 μL of Ex Taq polymerase (TaKaRa), and 2 μL of DNA template. The PCR amplifications were performed in a Bio-Rad iCycler (Bio-Rad, Hercules, CA, USA) as follows: pre-denaturation at 94°C for 4 min; 37 cycles at 94°C for 20 s, 56°C for 30 s, and 72°C for 50 s; and a final extension at 72°C for 5 min. The rbcL reaction mixture contained 34 μL of sterile distilled water, 2.5 μL of each primer, 10 μL of Pfu Plus 5× PCR Master Mix (ELPISBIO, Daejeon, Korea), and 1 μL of DNA template. The PCR amplifications were performed in a Bio-Rad T100 Thermal cycler (Bio-Rad) as follows: pre-denaturation at 95°C for 3 min; 37 cycles at 95°C for 20 s, 56°C for 20 s, and 72°C for 30 s; and a final extension at 72°C for 7 min.
The PCR products were analyzed on a 1% agarose gel using electrophoresis, and the target strips were observed under ultraviolet light. The products were sequenced by Bionics (Seoul, Korea).
Phylogenetic analysisThe sequences were viewed and analyzed using the Snap Gene Viewer (San Diego, CA, USA). Multiple sequence alignments between the target sequences and other related sequences of the SSU and rbcL genes obtained from the National Center for Biotechnology Information (NCBI) database (Supplementary Table S1) were performed using ClustalW (Thompson et al. 1994) in MEGA11. A total of 1,000 bootstrap replicates were calculated in the maximum likelihood (ML) tree construction for phylogenetic tree assessment. The Kimura 2-parameter model (Kumar et al. 2018) was selected as the nucleotide substitution model to estimate genetic differences and phylogenetic relationships.
Determination of biomass and productivityThe algal suspension was filtered through pre-weighted dry filters (1.2 μm Isopore Membrane filter GTTP04700; Millipore, Burlington, MA, USA). The filters were dried in an oven at 65°C overnight.
The dry weights were recorded on days 0, 3, 5, and 7. Biomass was used to determine average productivity (Pa) using the following equation:
, where Pa is the biomass productivity, dx is the harvest day, mo is the dry weight on day 0, and mx is the dry weight on the harvest day.
Pigment analysisPigment concentrations were measured using high performance liquid chromatography (HPLC) analysis (Baek et al. 2016). The cell suspension was centrifuged, and the supernatant was removed. Briefly, 1 mL of methanol was used to extract pigments from the samples. The methanol mixture was vortexed for 30 s and kept in a dark room at −20°C for 30 min. The samples were centrifuged at 4°C to obtain the supernatant. 500 μL supernatant was filtered using 0.2 μm HPLC syringe filters (Whatman, Buckinghamshire, UK).
HPLC analysis was performed using a Shimadzu HPLC LC-20AD Prominence (Shimadzu, Kyoto, Japan) equipped with a Spherisorb 5.0 μm ODS1 4.6 × 250 mm cartridge column (Waters, Milford, MA, USA). The pigment was separated using a solvent mixture containing 14% 0.1 M Tris-HCl, 84% acetonitrile, and 2% methanol from 0 to 15 min. From 15 to 19 min, the solvent mixture comprised 68% methanol and 32% acetonitrile. A post-run using the initial solvent mixture was conducted for 6 min. The flow rate was constant at 1.2 mL min−1. The pigments were detected at λ = 445 and 670 nm. The concentrations of individual pigments were determined from the HPLC profiles, which were calibrated using chlorophyll and carotenoid standards (14C Centralen; DHI, Horsholm, Denmark).
Fucoxanthin productivity (Pb) of N. palea HY1 was determined using HPLC analysis and previously obtained fucoxanthin concentrations using the following equation:
, where Pb is the fucoxanthin productivity, dx is the harvest day, Co is the concentration of fucoxanthin on 0 day, and Cx is the concentration of fucoxanthin on the harvest day.
RESULTSThe morphology of new freshwater diatom visualized by optical microscopy and SEM
Nitzschia palea, previously studied, was isolated from diverse freshwater environments worldwide and was found to present consistent morphological traits (Trobajo et al. 2009). Among predominant strains, the valves then exhibited a linear-to-lanceolate phenotype, featuring apices spanning the rostrate to the subcapitate. In most cases, the margins were parallel to the central zone. SEM analysis revealed that the striae ran uniseriately along the valve surface and adjacent to the raphe canal. In Fig. 1, light microscopy and SEM were used to examine the newly isolated freshwater diatoms for morphological identification. Bright-field and fluorescence images showed pennate diatoms, which were isopolar, elongate, and bilaterally symmetric in the isolated strain (Fig. 1A). The diatoms were solitary and linear-elliptical, with slightly drawn-out ends. Images of two plate-like plastids of new diatoms obtained using fluorescence microscopy revealed that the cells had two identical forms of chloroplasts (Fig. 1B & C).
Further observation by SEM images showed more detailed silica structures of freshwater diatom, including the length and width of valves, which were approximately 12.09 ± 1.06 and 4.05 ± 0.07 μm (n = 12 specimens), respectively (Fig. 1D). In addition, this strain possessed a slit along the long axis called the raphe, which is visible at the margin of the valve in the SEM (Fig. 1D–F). The diatoms portrayed marginal fibulae and transapical striae (Fig. 1F). Transapical striae were parallel in the middle, radiated slightly towards the apices, and finally converged at the apices (Fig. 1D). Poroid areolae are diatom pore areas that were arranged in a row with a diameter of approximately 56 ± 23 nm (Fig. 1D & E). Our observations suggest that the newly isolated freshwater diatom shares many morphologies with N. palea (Trobajo et al. 2006, 2009, Crowell et al. 2019).
Molecular phylogenetic analysis of Nitzschia palea HY1Morphological features clearly evidenced that the HY1 strain belongs to N. palea, however, to further confirm the identity of N. palea isolated in our study, the molecular analysis to determine the phylogenetic positions of N. palea HY1 was performed using SSU rRNA (Fig. 2A) and rbcL gene sequences (Fig. 2B). The primers used for the amplification and sequencing of nuclear SSU rRNA and rbcL are listed (Table 1). The SSU rRNA sequence of N. palea HY1 was aligned with the published sequences of other diatoms (two Nitzschia palea, and five Nitzschia sp., two Pseudo-nitzschia sp., Tryblionella apiculate, one Cylindrotheca sp., Phaeodactylum tricornutum, Navicula radiosa, and one dinoflagellate, Peridinium foliaceum) (Supplementary Table S1). The rbcL sequences of N. palea HY1 were also aligned with published sequences of other diatoms (two N. palea, six Nitzschia sp., and other diatoms: as Amphora berolinensis, Plagiogrammopsis castigates, Tryblionella debilis, Pseudo-nitzschia pungens, Phaeodactylum tricornutum, and Plagiogrammopsis castigates) (Supplementary Table S1). The ML phylogenetic analysis of SSU rRNA and rbcL genes was performed to establish the relationship of the new strain, “HY1” grouped into the clade with N. palea strains identified, indicating that HY1 was classified as N. palea (Fig. 2).
Biomass productivity of Nitzschia palea HY1Initially, we attempted to optimize the culture conditions using different culture media applied to freshwater diatoms, as previously reported (Beakes et al. 1988, Gérin et al. 2020). The growth curve of N. palea HY1 cultured in conventional DM (Beakes et al. 1988) was very slow (Supplementary Fig. S1). To improve the growth rate and increase biomass productivity, we used FDM, which has been previously studied (Gérin et al. 2020). Compared with cell growth in DM, N. palea HY1 showed a significant increase in growth for up to 7 days of cultivation in FDM before reaching a plateau (Supplementary Fig. S1). We then modified FDM by increasing the bicarbonate concentration from 2 to 10 mM, resulting in a strong increase in cell growth in the 10 mM bicarbonate-enriched FDM (which we called modified FDM [MFDM]) (Supplementary Table S2).
As shown in Supplementary Table S2, the primary nutrients, N, P, and K, were notably enriched in MFDM, and the inorganic carbon concentration increased by nearly fifty-fold compared to that in DM. The ample carbon supply provided by MFDM in this study resulted in a reduced time frame for reaching the maximum growth (Fig. 3). The initial biomass contents of N. palea HY1 were approximately 0.27 g L−1, as measured from cells cultured in both FDM, and MFDM, and the cultures were cultivated for 7 days. When cultivated in FDM, the maximum biomass content was 0.68 ± 0.06 g L−1 (Fig. 3A), and the maximum biomass productivity was calculated as 0.06 ± 0.03 mg L−1 d−1 (Fig. 3B). In contrast, using MFDM, the maximum biomass content was 1.83 ± 0.10 g L−1, and the maximum biomass productivity was 0.30 ± 0.08 g L−1 d−1. Therefore, increasing the bicarbonate concentration in the culture media resulted in 2.7-fold increase in the maximum biomass content and a five-fold increase in the maximum biomass productivity. Furthermore, we employed aeration at a specific speed (80 mL min−1) in MFDM, which led to an increase in the biomass (3.3 fold) compared to the control (shaking culture with MFDM at 130 rpm) with remarkably higher biomass productivity (Table 2, Fig. 4B).
Enhanced fucoxanthin production and productivity in Nitzschia palea HY1 cultured in MFDMTo measure the fucoxanthin content and volumetric productivity of N. palea HY1, the cells were cultured using the same growth conditions employed for the biomass content and productivity measurements. The fucoxanthin content and volumetric productivity of N. palea HY1 cultured in FDM and MFDM were compared in Fig. 5. The maximum fucoxanthin content was 9.19 ± 0.69 mg L−1 on day 7, with a maximum volumetric productivity of fucoxanthin (1.15 ± 0.12 mg L−1 d−1) in FDM (Fig. 5). However, in MFDM, the maximum fucoxanthin content was 9.15 ± 0.42 mg L−1 on day 5, and the maximum fucoxanthin productivity was calculated as 2.00 ± 0.35 mg L−1 d−1 on day 3. The results indicated that although the maximum fucoxanthin content was similar to the increasing concentration of 10 mM bicarbonate, the maximum productivity of fucoxanthin was approximately two-fold higher than that of cells cultured in FDM. Interestingly, further optimized MFDM with aeration enhanced the fucoxanthin concentration and productivity to 37.44 ± 2.98 mg L−1 and 5.10 ± 0.48 mg L−1 d−1, respectively (Table 2, Fig. 6).
DISCUSSIONIdentification of newly isolated freshwater diatomDiatoms are one of the most interesting microalgae for producing biomass and valuable commercial products, owing to their versatile roles and distinctive biomolecules to be applied (Marella et al. 2020). In this study, the initial morphological characteristics of newly isolated Korean freshwater diatoms were determined using light microscopy and SEM. As shown in Fig. 1, the valves of HY1 strain had a mean length of 12.09 ± 1.06 μm and a mean width of 4.05 ± 0.07 μm which are similar to most pennate diatoms (Trobajo et al. 2009). However, the reported length of N. palea cells has a wide range of 12–44 μm depending on the strains isolated worldwide (Trobajo et al. 2009). The N. palea strain isolated in the United States was identified and named “N. palea Wise.” This strain displays a distinctive feature: a series of fibulae that became evident along a single edge. SEM analysis revealed that the valves were linear-lanceolate to lanceolate. The average length of the valves of Wise was 25.8 μm, whereas the average width was 4.1 μm. The striae exhibited a parallel alignment, comprising uniseriate sequences of poroid areolae varying in shape from circular to rectangular. Furthermore, these striae extended transapically (Crowell et al. 2019). Strain Wise had the most common morphological features with strain HY1 except for the mean length because strain Wise was longer than strain HY1 (12.09 μm).
Furthermore, the taxonomic identification of the newly isolated N. palea HY1 was clearly confirmed by comparing its 18S rDNA region and rbcL with those of other N. palea strains that showed high similarity (Fig. 2A & B).
Biomass and fucoxanthin production of Nitzschia palea HY1Maximum productivity refers to the highest yield that can be achieved within a given set of times under the same culture conditions. Hence, the maximum productivity shown in the Figures and Table 2 represents the point at which each culture system operates at its peak performance, producing the greatest amount of the desired algal biomass and fucoxanthin output. Typically, biomass productivity is calculated based on the amount of biomass produced per unit of time. As shown in Fig. 3B, the productivity of N. palea biomass gradually decreased as the biomass increased. Notably, cultivating the HY1 strain in MFDM on day 3 resulted in maximum productivity. However, the algal biomass continued to increase and reached its maximum content on day 5. The highest yield of the desired algal biomass was observed on day 3 of culturing N. palea, which effectively minimized waste, optimized resource utilization, and saved time.
The majority of Nitzschia strains are typically benthic diatoms that reside at the bottom of water bodies and attach to substrates (Guo et al. 2022). However, in this study, we investigated a pinnate benthic strain, N. palea HY1, which displayed the ability to be cultured planktonically with sufficient turbulence. Interestingly, we sourced this strain from the surface of stones in a stream. Notably, N. palea HY1 has a natural tendency to aggregate, necessitating appropriate agitation or aeration for optimal growth. Therefore, by incorporating aeration in the MFDM culture medium, we observed a remarkable increase in the maximum biomass and volumetric productivity compared to cultures without aeration (Table 2, Fig. 4).
Fucoxanthin production may vary considerably depending on the species. It is essential to adopt successful culturing approaches for biomass and fucoxanthin production to ensure sustainability, feasibility, and economic viability. Fucoxanthin production has been reported to vary under many nutrients, such as nitrogen, phosphate, silicate, and inorganic carbon, or light for the cultivation of diatoms (Lu et al. 2018). In a previous study, the amount of fucoxanthin present in biomass remained highly consistent in FDM, with 5.5 mg g−1 dry cell weight (DCW) being the highest yield (Gérin et al. 2020). However, HY1 strain in this study had a yield of 13.7 mg g−1 DCW in the same media, which is 2.5-fold higher than the reported strain (Table 2) (Gérin et al. 2020). Furthermore, the utilization of MFDM in our research resulted in a reduced cultivation duration, with a maximum of 5 days required to achieve maximum biomass (Fig. 5). This difference is likely due to the body size of Nitzschia strains; the length of Nitzschia used in the previous study was much longer (~20 μm) (Gérin et al. 2020) than that of HY1 (~12 μm) (Fig. 1). According to Litchman et al. (2009), body size is a fundamental characteristic of organisms that influences nearly all aspects of their physiology.
Seth et al. (2021) have stated that the effective generation of biomass and fucoxanthin from diatoms has faced various challenges. Nevertheless, these barriers can be overcome by improving the culture conditions or discovering new strains possessing the desired characteristics. Hence, we characterized a new freshwater diatom, the N. palea HY1 strain, and modified FDM by adding bicarbonate to enhance biomass and fucoxanthin content. Consequently, the biomass grown in MFDM was 2.7 times higher than that in the original FDM, with an increase in biomass productivity (Table 2, Fig. 3).
The fucoxanthin content of most species ranges from 1 to 10 mg g−1 of dry biomass. However, some marine species accumulate more than 20 mg g−1 of fucoxanthin (Wang et al. 2021) with low biomass productivity. In addition, some marine strains (Nitzschia laevis and Phaeodactylum tricotunum) showing higher biomass and fucoxanthin yields than those of freshwater diatom, grew under mixotrophic conditions in the presence of glucose or glycerol as a carbon source (Table 2). However, under photoautotrophic condition, N. palea HY1 cultivated in optimized MFDM showed a maximum fucoxanthin productivity of 5.10 mg L−1 d−1 while the marine microalga P. tricotunum showed 4.74 mg L−1 d−1 (Table 2), which is slightly less than that of the N. palea HY1. So far, marine diatoms have received greater attention than freshwater diatoms because of their rapid growth rates and their ability to thrive in open-sea ponds along coastal areas (Khaw et al. 2022). Nevertheless, the cultivation of marine microalgae in photobioreactors can present challenges such as corrosion and crystallization, which are dependent on the type of bioreactor used. This necessitates an additional step of salt removal for the biomass. As a solution to these challenges, freshwater microalgae can be utilized as an alternative source of fucoxanthin production. Thus, we conducted a study to determine the necessary culture conditions for fucoxanthin production in freshwater diatoms.
Regarding fucoxanthin production, productivity was 2 mg L−1 d−1 without air bubbling. However, with air bubbling, it increased to 3 mg L−1 d−1 on day 3, and further to a maximum of 5 mg L−1 d−1 on day 5 (Fig. 6B). From this optimization, maximizing fucoxanthin productivity ensures that a larger quantity of fucoxanthin is produced using given amounts of resources (e.g., algal biomass, nutrients, and energy). This optimization may reduce the production cost per output unit, making the process more economically viable. This high productivity can allow fucoxanthin production to compete more effectively with previously described production methods. Higher productivity can facilitate scaling up of the production process, resulting in economies of scale and further reducing production costs.
In this study, we refined and optimized the culture media and conditions to enhance the biomass and fucoxanthin production of the benthic freshwater diatom. This study serves as an important milestone in the exploration of future applications for freshwater diatoms in research. Subsequent studies have the potential to unveil freshwater diatoms with considerably enhanced fucoxanthin productivity, surpassing the marine species and N. palea HY1 examined in our study.
ACKNOWLEDGEMENTSThis research was supported by Korea Environmental Industry & Technology Institute (KEITI) through “The Project to develop eco-friendly new materials and processing technology derived from wildlife,” funded by the Korea Ministry of Environment (MOE) (2021003270007).
SUPPLEMENTARY MATERIALS
Supplementary Table S1. Isolation information and GenBank accession number of diatoms use in this study (https://www.e-algae.org).
algae-2023-38-3-191-Supplementary-Table-S1.pdf
Supplementary Table S2. Comparison of nutrient composition between standard freshwater diatom media and modified FDM media (https://www.e-algae.org).
algae-2023-38-3-191-Supplementary-Table-S2.pdf
Supplementary Fig. S1. Growth curve of Nitzschia palea HY1 in various types of media such as diatom medium (DM, triangle), freshwater medium (FDM, square), and modified freshwater medium (MFDM, circle) (https://www.e-algae.org).
algae-2023-38-3-191-Supplementary-Fig-S1.pdf
Fig. 1Optical and scanning electron microscopic images of Nitzschia palea HY1. (A) Bright-field image. (B) Autofluorescence image. (C) Merged image of living cells. (D–F) Intact frustule of N. palea HY1 using scanning electron microscopy. (D) External view with the position of the canal raphe—the strongly hooked apical raphe endings. (E) Internal view of the presence of the small fibulae and pores in a row. (D & E) Exterior. Fibulae appear as a row of lighter spots along one margin. (F) Interior of a valve: specimen in internal view of the presence of the small fibulae and pores in a raw. Scale bars represent: A–C, 10 μm; D & E, 2 μm; F, 1 μm. 
Fig. 2Molecular phylogenetic analysis using the maximum likelihood (ML) method based on the small subunit ribosomal RNA (SSU rRNA) (A) and rbcL (B) genes showing the phylogenetic. The evolutionary history was inferred using the ML method and the Kimura 2-parameter model. The trees with the highest log likelihoods (−4,273.35 and −5,414.86 for SSU rRNA and rbcL, respectively) are shown. The percentage of trees in which the associated taxa clustered together is shown above the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood approach and then selecting the topology with superior log likelihood value. A discrete Gamma distribution was used to model evolutionary rate differences among sites (6 categories [+G, parameter = 0.2000]). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved 14 nucleotide sequences. There were a total of 1,497 positions in the final dataset. Evolutionary analyses were conducted in MEGA11. 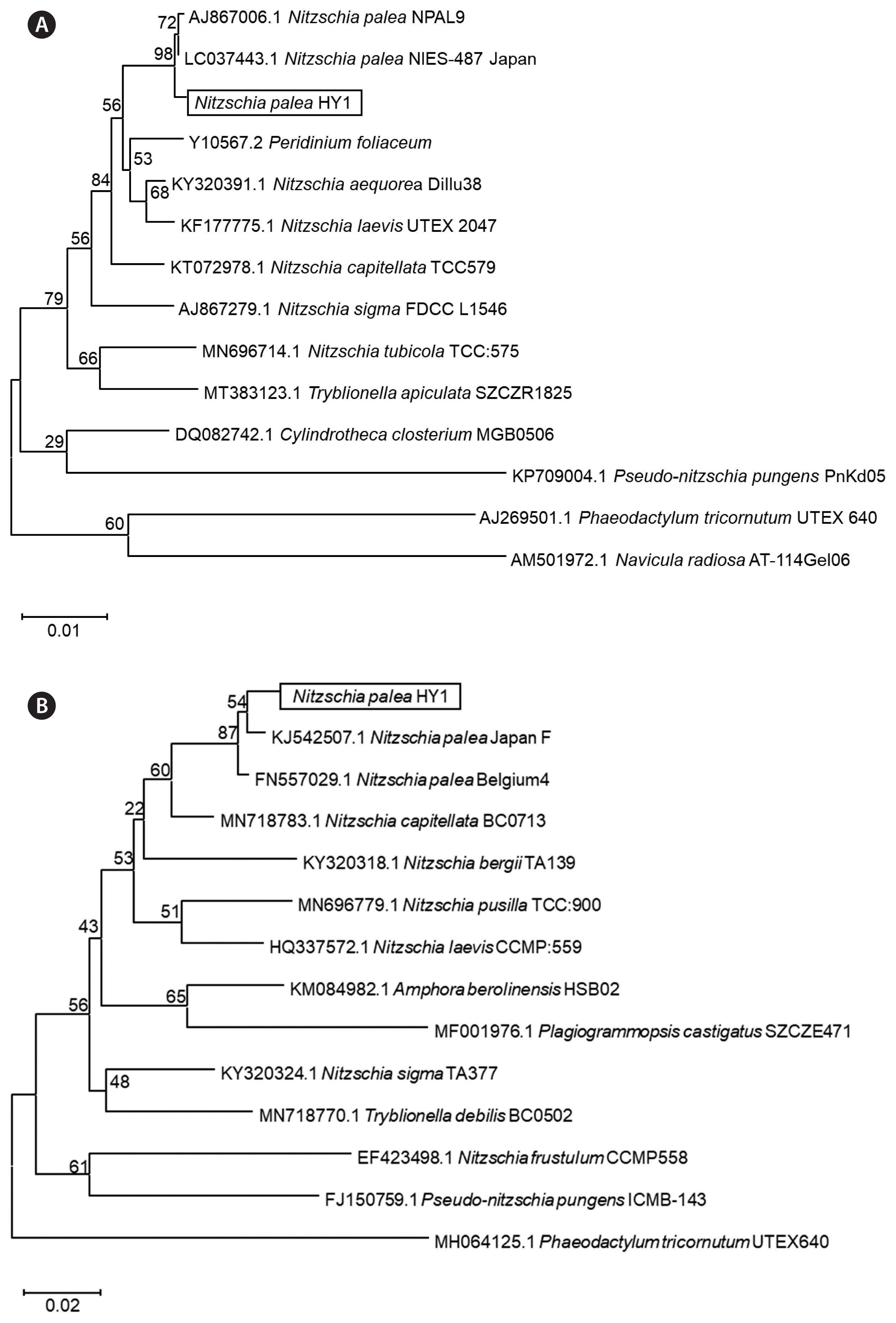
Fig. 3Biomass concentration (g L−1) (A) and biomass productivity (g L−1 d−1) (B) in Nitzschia palea HY1 cultivated in FDM (shaking at 130 rpm, open bar and open circle) and MFDM (shaking at 130 rpm, gray bar and open square). FDM, freshwater medium; MFDM, modified freshwater medium. All data represent means with standard deviations from at least three independent experiments. Statistical analysis was performed using Student’s t-test (***p < 0.001). 
Fig. 4Biomass concentration (g L−1) (A) and biomass productivity (g L−1 d−1) (B) in Nitzschia palea HY1 cultivated in MFDM (stirred at 500 rpm, open bar and open circle) and aerated MFDM (gray bar and open square). MFDM, modified freshwater medium. All data represent means with standard deviations from at least three independent experiments. Statistical analysis was performed using Student’s t-test (*p < 0.05, **p < 0.01). 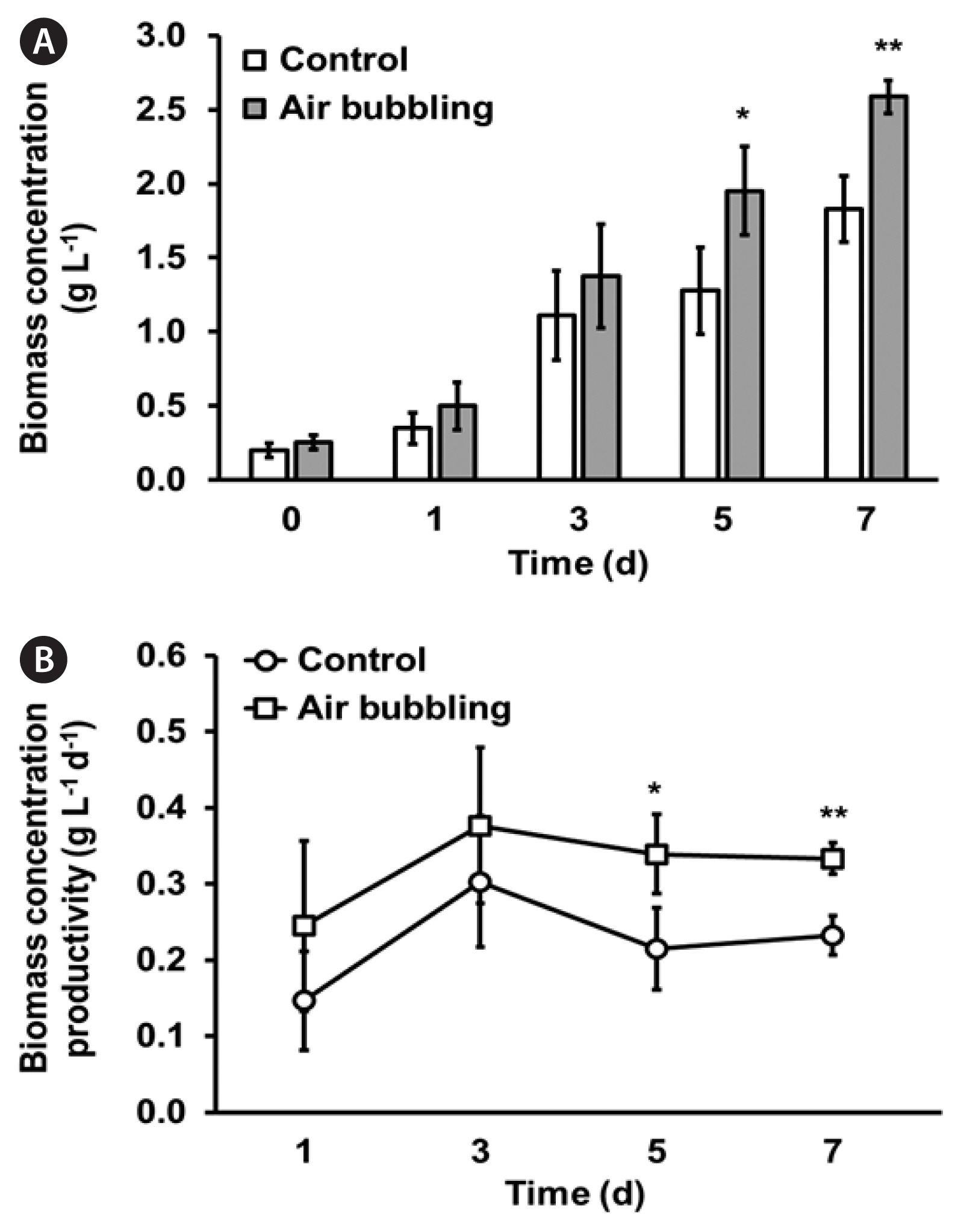
Fig. 5Fucoxanthin concentration (mg L−1) (A) and productivity of fucoxanthin (mg L−1 d−1) (B) in Nitzschia palea HY1 cultivated in FDM (shaking at 130 rpm, open bar and open circle) and MFDM (shaking at 130 rpm, gray bar and open square). FDM, freshwater media; MFDM, modified freshwater medium. All data represent means with standard deviations from at least three independent experiments. Statistical analysis was performed using Student’s t-test (*p < 0.05, **p < 0.01). 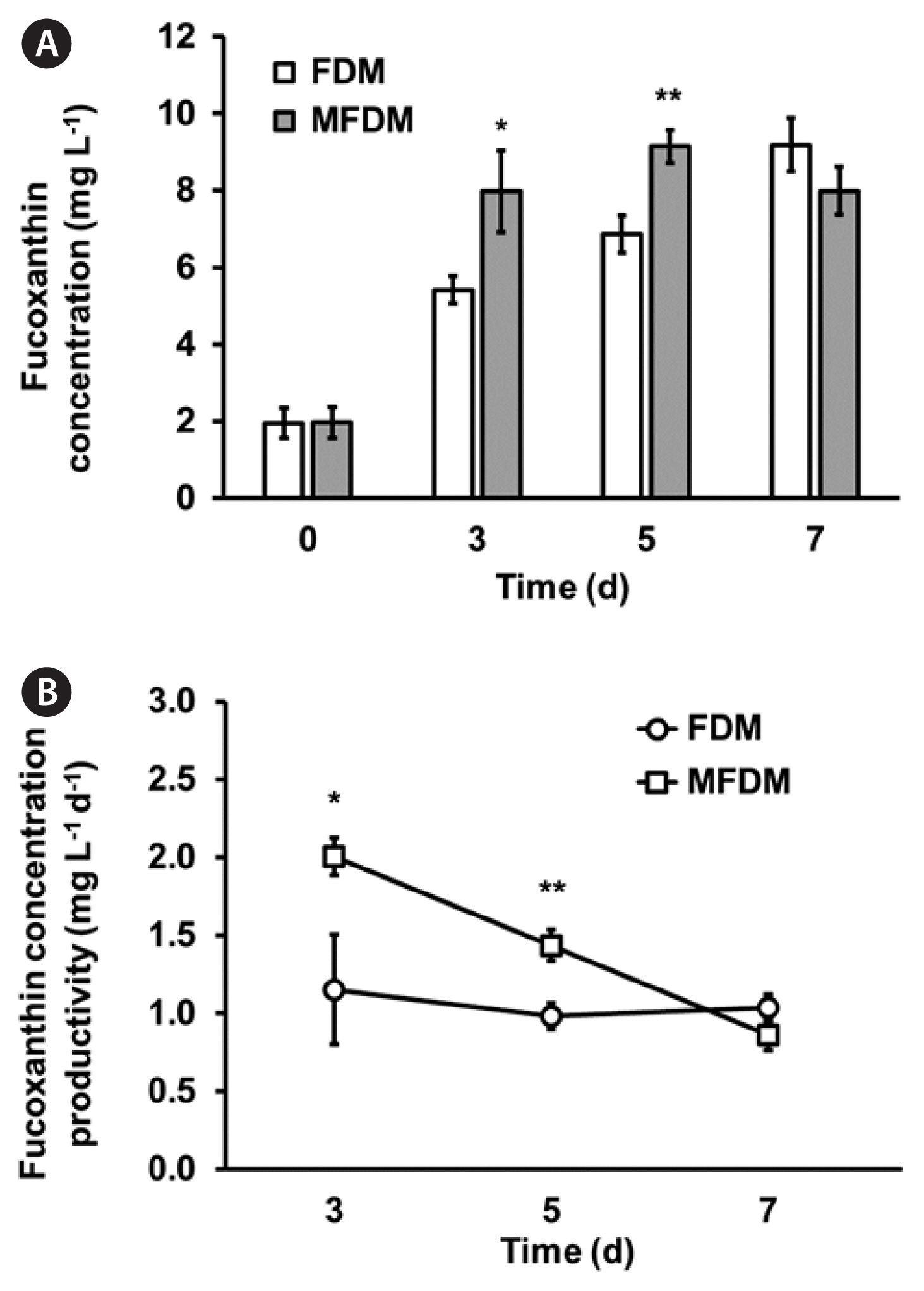
Fig. 6Fucoxanthin concentration (mg L−1) (A) and productivity of fucoxanthin (mg L−1 d−1) (B) in Nitzschia palea HY1 cultivated in MFDM (stirred at 500 rpm, open bar and open circle) and aerated MFDM (gray bar and open square). MFDM, modified freshwater medium; all data represent means with standard deviations from at least three independent experiments. Statistical analysis was performed using Student’s t-test (*p < 0.05, ***p < 0.001). 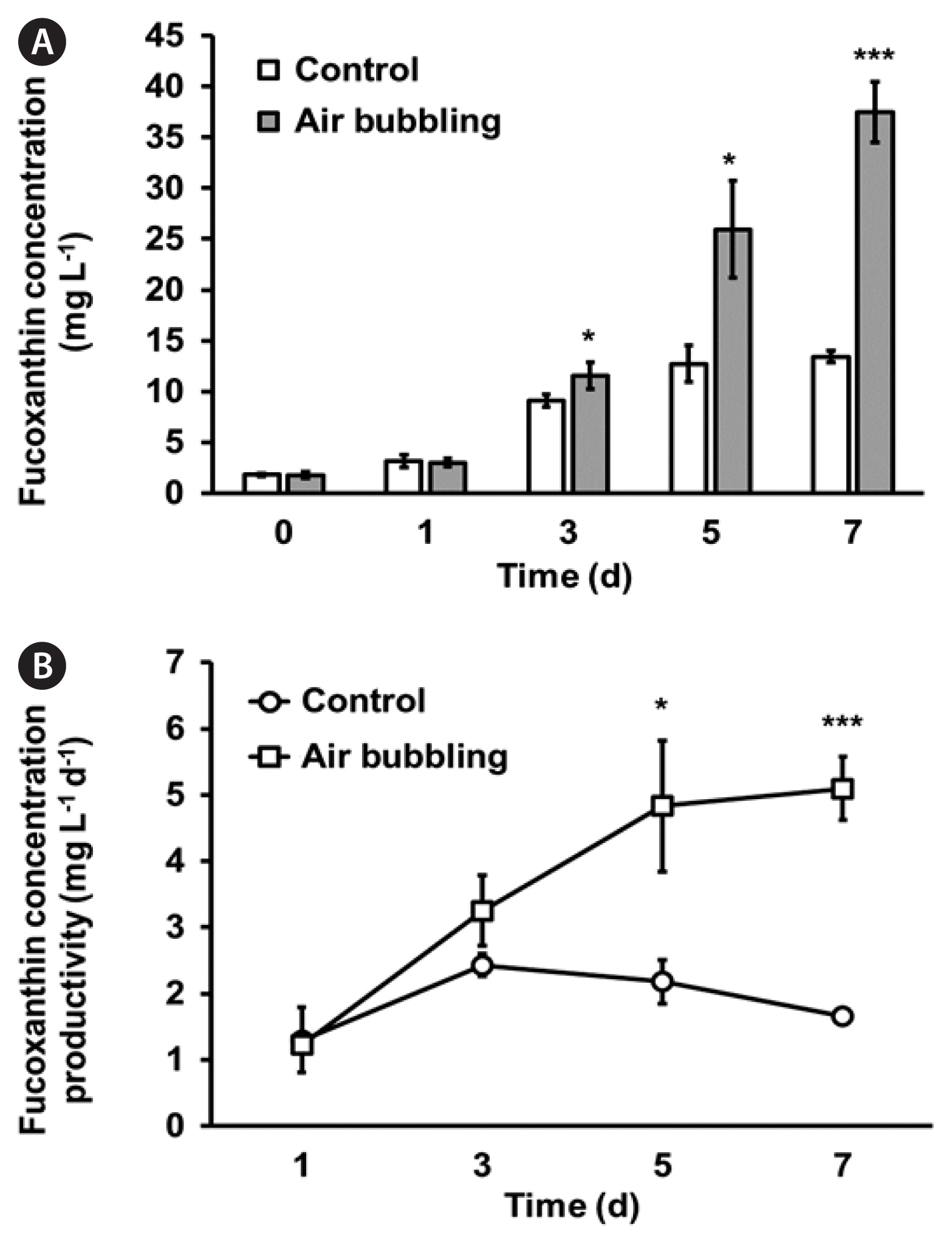
Table 1Primers used for amplification and sequencing of the nuclear SSU rRNA and rbcL gene
Table 2Biomass, fucoxanthin content and productivity from various diatoms in this study compared to the previous works of literature
FDM, freshwater media; MFDM, modified FDM; MCOMBO, modified COMBO (Kilham et al. 1998); LDM, Lewin’s marine diatom medium; L : D, light and dark; R : B, red and blue; D (#), days of cultivation. REFERENCESAndersen, R. A. & Kawachi, M. 2005. Traditional microalgae isolation techniques. In : Anderson R. A., editor Algal Culturing Techniques. Elsevier Academic Press, Burlington, MA, 83–92.
Baek, K., Kim, D. H., Jeong, J., Sim, S. J., Melis, A., Kim, J.-S., Jin, E. & Bae, S. 2016. DNA-free two-gene knockout in Chlamydomonas reinhardtii via CRISPR-Cas9 ribonucleoproteins. Sci. Rep. 6:30620 pp.
Beakes, G. W., Canter, H. M. & Jaworski, G. H. M. 1988. Zoospore ultrastructure of Zygorhizidium affluens and Z. planktonicum, two chytrids parasitizing the diatom Asterionella formosa
. Can. J. Bot. 66:1054–1067.
Bruder, K. & Medlin, L. K. 2007. Molecular assessment of phylogenetic relationships in selected species/genera in the naviculoid diatoms (Bacillariophyta). I. The genus Placoneis
. Nova Hedwigia. 85:331–352.
Chonova, T., Kurmayer, R., Rimet, F., Labanowski, J., Vasselon, V., Keck, F., Illmer, P. & Bouchez, A. 2019. Benthic diatom communities in an alpine river impacted by waste water treatment effluents as revealed using DNA metabarcoding. Front. Microbiol. 10:653 pp.
Crowell, R. M., Nienow, J. A. & Cahoon, A. B. 2019. The complete chloroplast and mitochondrial genomes of the diatom Nitzschia palea (Bacillariophyceae) demonstrate high sequence similarity to the endosymbiont organelles of the dinotom Durinskia baltica
. J. Phycol. 55:352–364.
Finlay, B. J., Monaghan, E. B. & Maberly, S. C. 2002. Hypothesis: the rate and scale of dispersal of freshwater diatom species is a function of their global abundance. Protist. 153:261–273.
Gao, B., Chen, A., Zhang, W., Li, A. & Zhang, C. 2017. Co-production of lipids, eicosapentaenoic acid, fucoxanthin, and chrysolaminarin by Phaeodactylum tricornutum cultured in a flat-plate photobioreactor under varying nitrogen conditions. J. Ocean Univ. China. 16:916–924.
Gérin, S., Delhez, T., Corato, A., Remacle, C. & Franck, F. 2020. A novel culture medium for freshwater diatoms promotes efficient photoautotrophic batch production of biomass, fucoxanthin, and eicosapentaenoic acid. J. Appl. Phycol. 32:1581–1596.
Griffiths, M. J. & Harrison, S. T. L. 2009. Lipid productivity as a key characteristic for choosing algal species for biodiesel production. J. Appl. Phycol. 21:493–507.
Guo, X., Tang, Y., Yin, J., Li, R., Qin, B., Jiang, L., Chen, X. & Huang, Z. 2022. Long-term manganese exposure-mediated benthic diatom assemblage in a subtropical stream: distribution, substrate preferences and Mn-tolerance. J. Environ. Manag. 322:116153 pp.
Jeong, B.-R., Jang, J. & Jin, E. 2023. Genome engineering via gene editing technologies in microalgae. Bioresour. Technol. 373:128701 pp.
Kanazawa, K., Ozaki, Y., Hashimoto, T., Das, S. K., Matsushita, S., Hirano, M., Okada, T., Komoto, A., Mori, N. & Nakatsuka, M. 2008. Commercial-scale preparation of biofunctional fucoxanthin from waste parts of brown sea algae Laminalia japonica.
. Food Sci. Technol. Res. 14:573–582.
Khaw, Y. S., Yusoff, F. M., Tan, H. T., Noor Mazli, N. A. I., Nazarudin, M. F., Shaharuddin, N. A., Omar, A. R. & Takahashi, K. 2022. Fucoxanthin production of microalgae under different culture factors: a systematic review. Mar. Drugs. 20:592 pp.
Ki, J. S. & Han, M.-S. 2005. Molecular analysis of complete SSU to LSU rDNA sequence in the harmful dinoflagellate Alexandrium tamarense (Korean isolate, HY970328M). Ocean Sci. J. 40:43–54.
Kilham, P. & Hecky, R. E. 1988. Comparative ecology of marine and freshwater phytoplankton. Limnol. Oceanogr. 33:776–795.
Kilham, S. S., Kreeger, D. A., Lynn, S. G., Goulden, C. E. & Herrera, L. 1998. COMBO: a defined freshwater culture medium for algae and zooplankton. Hydrobiologia. 377:147–159.
Kim, S. M., Kang, S.-W., Kwon, O.-N., Chung, D. & Pan, C.-H. 2012. Fucoxanthin as a major carotenoid in Isochrysis aff. galbana: characterization of extraction for commercial application. J. Korean Soc. Appl. Biol. Chem. 55:477–483.
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35:1547–1549.
Litchman, E., Klausmeier, C. A. & Yoshiyama, K. 2009. Contrasting size evolution in marine and freshwater diatoms. Proc. Natl. Acad. Sci. U. S. A. 106:2665–2670.
Lu, X., Sun, H., Zhao, W., Cheng, K.-W., Chen, F. & Liu, B. 2018. A hetero-photoautotrophic two-stage cultivation process for production of fucoxanthin by the marine diatom Nitzschia laevis. Mar. Drugs. 16:219 pp.
Maberly, S. C., Gontero, B., Puppo, C., Villain, A., Severi, I. & Giordano, M. 2021. Inorganic carbon uptake in a freshwater diatom, Asterionella formosa (Bacillariophyceae): from ecology to genomics. Phycologia. 60:427–438.
Marella, T. K., López-Pacheco, I. Y., Parra-Saldívar, R., Dixit, S. & Tiwari, A. 2020. Wealth from waste: diatoms as tools for phycoremediation of wastewater and for obtaining value from the biomass. Sci. Total Environ. 724:137960 pp.
Nelson, D. M., Tréguer, P., Brzezinski, M. A., Leynaert, A. & Quéguiner, B. 1995. Production and dissolution of biogenic silica in the ocean: revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Glob. Biogeochem Cycles. 9:359–372.
Seth, K., Kumar, A., Rastogi, R. P., Meena, M., Vinayak, V. & Harish, 2021. Bioprospecting of fucoxanthin from diatoms: challenges and perspectives. Algal Res. 60:102475 pp.
Thompson, J. D., Higgins, D. G. & Gibson, T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680.
Tiam, S. K., Lavoie, I., Doose, C., Hamilton, P. B. & Fortin, C. 2018. Morphological, physiological and molecular responses of Nitzschia palea under cadmium stress. Ecotoxicology. 27:675–688.
Tréguer, P., Bowler, C., Moriceau, B., Dutkiewicz, S., Gehlen, M., Aumont, O., Bittner, L., Dugdale, R., Finkel, Z., Iudicone, D., Jahn, O., Guidi, L., Lasbleiz, M., Leblanc, K., Levy, M. & Pondaven, P. 2018. Influence of diatom diversity on the ocean biological carbon pump. Nat. Geosci. 11:27–37.
Trobajo, R., Clavero, E., Chepurnov, V. A., Sabbe, K., Mann, D. G., Ishihara, S. & Cox, E. J. 2009. Morphological, genetic and mating diversity within the widespread bioindicator Nitzschia palea (Bacillariophyceae). Phycologia. 48:443–459.
Trobajo, R., Mann, D. G., Chepurnov, V. A., Clavero, E. & Cox, E. J. 2006. Taxonomy, life cycle, and auxosporulation of Nitzschia fonticola (Bacillariophyta). J. Phycol. 42:1353–1372.
Vilmi, A., Karjalainen, S. M., Landeiro, V. L. & Heino, J. 2015. Freshwater diatoms as environmental indicators: evaluating the effects of eutrophication using species morphology and biological indices. Environ. Monit. Assess. 187:243 pp.
Wang, S., Wu, S., Yang, G., Pan, K., Wang, L. & Hu, Z. 2021. A review on the progress challenges and prospects in commercializing microalgal fucoxanthin. Biotechnol. Adv. 53:107865 pp.
Xia, S., Wang, K., Wan, L., Li, A., Hu, Q. & Zhang, C. 2013. Production, characterization, and antioxidant activity of fucoxanthin from the marine diatom Odontella aurita. Mar. Drugs. 11:2667–2681.
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||