ABSTRACT
Cochlodinium polykrikoides is a red-tide forming dinoflagellate that causes significant worldwide impacts on aquaculture industries and the marine ecosystem. There have been extensive studies on managing and preventing C. polykrikoides blooms, but it has been difficult to identify an effective method to control the bloom development. There is also limited genome information on the molecular mechanisms involved in its various ecophysiology and metabolism processes. Thus, comprehensive genome information is required to better understand harmful algal blooms caused by C. polykrikoides. We estimated the C. polykrikoides genome size using flow cytometry, with detection of the fluorescence of DNA stained with propidium iodide (PI). The nuclear genome size of C. polykrikoides was 100.97 Gb, as calculated by comparing its mean fluorescence intensity (MFI) to the MFI of Mus musculus, which is 2.8 Gb. The exceptionally large genome size of C. polykrikoides might indicate its complex physiological and metabolic characteristics. Our optimized protocol for estimating the nuclear genome size of a dinoflagellate using flow cytometry with PI can be applied in studies of other marine organisms.
INTRODUCTIONThe nuclear genome size of eukaryotes (or protists) has been estimated to be up to one million-folds (Gregory et al. 2007). Intuitively, it is expected that more complex organisms have larger genomes, but there is a lack of correlation between eukaryotic genome size and its organismic complexity, which is called the C-value paradox (Cavalier-Smith 2005, Elliott and Gregory 2015). Instead, other possible correlations have been suggested. For example, organism-level traits (such as cell size, growth rate, population size, and metabolism) are correlated with genome size, but not genome-level traits (such as gene content, transposable element content, and related features) (Gregory 2001, 2002, Lynch and Conery 2003, Vinogradov 2004, Beaulieu et al. 2008, Gregory and Witt 2008). The highly compacted small genomes for which data can be obtained easily have received attention for the study of the evolutionary forces driving nuclear genome miniaturization and expansion (Cavalier-Smith 2005). In addition, extremely large genomes have been studied, but many genomic aspects remain unclear.
Over the last few decades, most coastal regions worldwide have been affected by harmful algal blooms (HABs). These phenomena are caused by blooms of algae, including dinoflagellates, diatoms, and seaweeds. These have had adverse impacts on human health, living marine resources, marine ecosystems, and recreational use of coastal areas (Zingone and Enevoldsen 2000, Anderson et al. 2012, Kang et al. 2014). As unicellular eukaryotes, dinoflagellates have a small cell size of 10–100 μm, but their nuclear genome size varies from 1 to 270 Gb, indicating a size that is one-third to 90-fold the size of the human genome (Lin 2011, Wisecaver and Hackett 2011). Recently, two dinoflagellates (Symbiodinium minutum and Symbiodinium kawagutii), which have relatively small genome sizes (1.5 and 1.2 Gb, respectively), were sequenced, revealing their unique and divergent genomic characteristics (Shoguchi et al. 2013, Lin et al. 2015).
Among the roughly 300 toxic and nontoxic microalgal species involved in HABs, Cochlodinium polykrikoides Margalef has been considered one of the primary species affecting the marine ecosystem and fishing industry. It causes mass mortalities of wild and farmed fish, with severe impacts on fish aquaculture and economic damage (Dorantes-Aranda et al. 2010, Gárate-Lizárraga 2013, Wolny et al. 2015, Guo et al. 2016). Recently, there have been frequent C. polykrikoides blooms in coastal areas of the eastern and western Pacific Ocean, western Atlantic Ocean, and Indian Ocean (Curtiss et al. 2008, Gobler et al. 2008, Richlen et al. 2010). For management of coastal resources and protection of public health, it is necessary for the molecular mechanisms of the C. polykrikoides blooms to be fully understood at the genome level. Thus, a genome size estimation is an important initial step in building the genome-wide knowledge of C. polykrikoides.
In this study, we first investigated the C. polykrikoides nuclear genome size using flow cytometry with propidium iodide (PI) because the genome size may provide further insights into the functional complexity of its physiological and metabolic characteristics, which can be used to prevent or reduce its harmful blooms.
MATERIALS AND METHODSCulture of Cochlodinium polykrikoides
C. polykrikoides was cultured in autoclaved seawater enriched with the f/2 medium at 23 ± 1°C and a salinity of 32 psu (Guillard and Ryther 1962). The light was provided by four 36-W daylight fluorescent lamps (Dulux L 36W/865; Osram, Münich, Germany) and maintained at 300 μmol photons m−2 s−1 and a 12 : 12 LD cycle. Fresh f/2 medium was added every 4–5 d. Unless otherwise noted, all chemicals used in the study were purchased from Sigma-Aldrich Co. (St. Louis, MO, USA).
Propidium iodide staining for flow cytometry analysisCultured C. polykrikoides (approximately 4 × 105 cells) were harvested by centrifugation at 1,000 ×g for 5 min at 4°C and then washed in a methanol : acetic acid (3 : 1) solution for 20 min on ice twice to eliminate chlorophyll, which affects fluorescence intensity during flow cytometry analysis. After fixation in 90% methanol for 5 min on ice, cells were resuspended in 20 mM 3-(N-morpholino) propanesulfonic acid, 4-morpholinepropanesulfonic acid (MOPS)-buffer, pH 7.2 (LaJeunesse et al. 2005).
Mouse thymuses were dissected from euthanized ICR mice following cervical dislocation and kept on Dulbecco’s modified Eagle’s medium with 1% penicillin and streptomycin, 44.5 mM sodium bicarbonate, and 10% fetal bovine serum. Single thymus cells were separated by passing the mixture through a 70-μm cell strainer in 1 mL of phosphate buffered saline (PBS) after blood cells from the thymus were washed off with PBS (pH 7.2). After fixation in 70% ethanol for 2 h at 4°C, 5 × 106 thymus cells were resuspended in PBS for PI staining. Then, 0.1 mg mL−1 RNase A (20 mg mL−1; Invitrogen, Carlsbad, CA, USA) was added to eliminate the non-specific PI signals from RNA in the PI staining solution (50 μg mL−1 of PI in PBS). PI nuclear DNA staining of C. polykrikoides and mouse thymus cells was conducted for 20 minutes at room temperature in the dark (LaJeunesse et al. 2005).
Flow cytometry analysisPI-stained cells were analyzed with a BD FACSCalibur flow cytometer equipped with 14.5–15.5 mw laser power and a 488-nm laser and 670 LP filter options (BD Biosciences, San Diego, CA, USA). Data were acquired while running at a low flow rate of 6.30–6.50 V until 20,000 cells were analyzed and processed using WinMDI ver. 2.9 (The Scripps Research Institute, San Diego, CA, USA). To avoid disturbance of the fluorescence by debris and attachment of multiple cells, only single cells were gated, and the cytograms present forward scattered light (FSC) on the X-axis and side scattered light (SSC) on the Y-axis. Single-cell DNA fluorescence in G0/G1 phase is represented in the histogram as PI fluorescence intensity (FL3) relative to cell counts.
The value of the geometric log mean was used as the mean fluorescence intensity (MFI) for calculating the unknown genome size. The genome size of C. polykrikoides was calculated according to the following formula (Hare and Johnston 2011) using Mus musculus as a reference genomic length of 2.8 Gb (Gregory et al. 2002).
RESULTS AND DISCUSSIONAlthough differences in nuclear genome size have no clear relationship with the number of genes, known as the C-value paradox, it might be helpful to understand an organism at the molecular level of genetics. Prior to understanding the genetic architecture, we first estimated the genome size of C. polykrikoides using flow cytometry with PI, which intercalates into DNA with little or no sequence biases.
Single cells were used in the cytograms to calculate the MFI of M. musculus and C. polykrikoides (Fig. 1A & B). Gated cells are presented in a histogram. The difference in fluorescence intensity between the two organisms on a logarithmic scale indicated that the genome size of C. polykrikoides was considerably larger than that of M. musculus (Fig. 1C). The statistics of the gated PI-stained cells from the flow cytometry data are summarized in Table 1. The numbers of events were gated cells contained in each peak over the total acquired cells for analysis (% Total). The percentage of cells in the gated region (% Gated) reflected the rates of single cells over possible fluorescence noise due to the condition of the samples such as cell debris and fragmented DNAs.
Using MFIs (7 and 257) of the first replicate a, the nuclear genome size was estimated as 100.97 Gb, based on the known mouse genomic length of 2.8 Gb. Similarly, the calculated genome size of the second replicate b was 110.54 Gb. Even though there were differences between the two independent replicates in number and peak height of events, the estimated nuclear genome size of C. polykrikoides was not significantly different between the two replicates, indicating that the nuclear genome size estimate was accurate. Because the first replicate a had many more single isolated cells and lower differences between gated cells in peak events, it is more reliable for estimating the genome size.
In order to compare the genome size of C. polykrikoides with those of other dinoflagellates, the data for previously examined dinoflagellate species are plotted in Fig. 2 in ascending order of genome size (Veldhuis et al. 1997, LaJeunesse et al. 2005, Shoguchi et al. 2013, Lin et al. 2015). The estimated genome size of 26 dinoflagellate species ranged widely in the scatter plot from 1.18 Gb for the recently sequenced Symbiodinium kawagutii to 271.94 Gb for Procentrum micans; there were even some different genome sizes for a species as estimated by two DNA-specific staining fluorophores, PicoGreen and SYTOX green (Veldhuis et al. 1997).
The genome size of C. polykrikoides ranked the fifth largest among the 26 dinoflagellates (Fig. 2). High numbers of gene copies, repetitive sequences, or multiple copies of noncoding DNA elements such as pseudogenes can result in an extremely large genome (Thornhill et al. 2007, Wisecaver and Hackett 2011, Ebenezer and Ki 2014). Further investigation of the whole or partial genome sequence will unveil how and why C. polykrikoides has a large genome size.
CONCLUSIONWe first estimated the C. polykrikoides nuclear genome size by flow cytometry with PI and hypothesized that the exceptionally large genome size might be responsible for their complex physiological and metabolic characteristics. Furthermore, the study of this extraordinary feature of C. polykrikoides will help us to understand the molecular mechanisms of the recent worldwide blooms. This methodology using FACS with PI staining is necessary in applications to estimate the nuclear genome size of algae with autofluorescent plastid pigments before performing whole genome sequencing.
ACKNOWLEDGEMENTSThis work was supported by the programs “Genome Analysis of Marine Organisms and Development of Functional Applications (PJT200620)” funded by MOF to CP and “Management of Marine Organisms causing Ecological Disturbance and Harmful Effects” funded by KIMST/MOF and NRF-2016R1A6A1A03012647 to KYK.
Fig. 1Flow cytometric analysis of Mus musculus and Cochlodinium polykrikoides stained with propidium iodide (PI). Of the acquired cells, only 2,000 cells are represented on the graph. (A) Single cells in the cytograms of M. musculus and C. polykrikoides, with forward scattered light (FSC) on the X-axis and side scattered light (SSC) on the Y-axis. (B) The gated single cell populations and their fluorescence intensities are indicated with SSC on the X-axis and PI fluorescence intensity (FL3) on the Y-axis. (C) G0/G1 DNA peaks of two species are shown in the histogram with FL3 on the X-axis and cell counts (events) on the Y-axis. 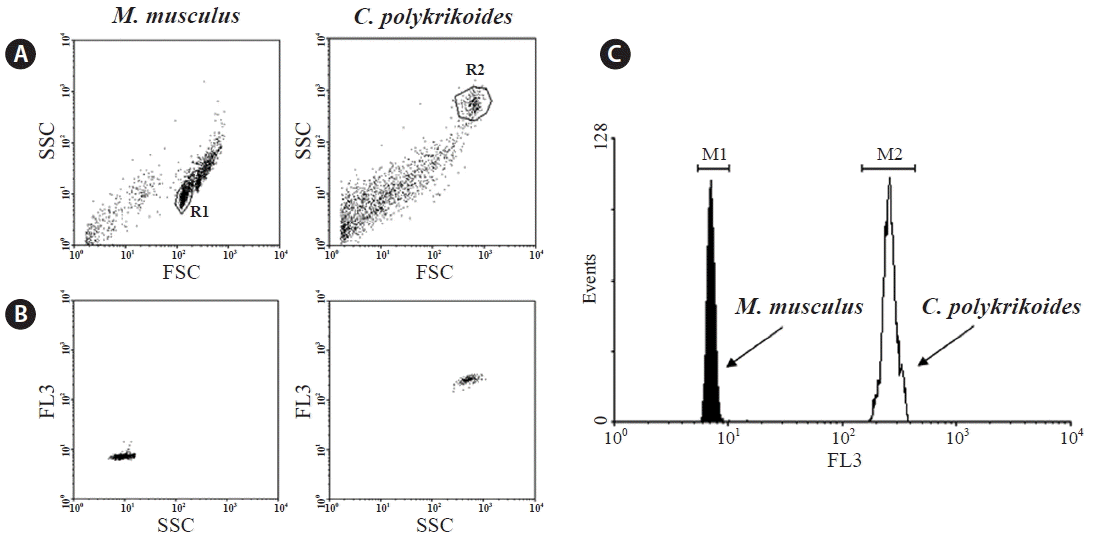
Fig. 2Genome size distribution of dinoflagellate species. Estimated genome sizes of 27 dinoflagellates ranged from 1.18 Gb to 271.94 Gb in the scatter plot. The genome size of Cochlodinium polykrikoides was positioned at 100.97 Gb, which was the fifth largest genome size. The genomic sizes of the 26 other dinoflagellates were obtained from Veldhuis et al. (1997), LaJeunesse et al. (2005), Shoguchi et al. (2013), and Lin et al. (2015). C-values were processed with a conversion factor, 0.978 Gb/pg (Dolezel et al. 2003). 
Table 1Summary statistics of the flow cytometry data in Fig. 1C
REFERENCESAnderson, DM., Cembella, AD. & Hallegraeff, GM. 2012. Progress in understanding harmful algal blooms: paradigm shifts and new technologies for research, monitoring, and management. Ann Rev Mar Sci. 4:143–176.
Beaulieu, JM., Leitch, IJ., Patel, S., Pendharkar, A. & Knight, CA. 2008. Genome size is a strong predictor of cell size and stomatal density in angiosperms. New Phytol. 179:975–986.
Cavalier-Smith, T. 2005. Economy, speed and size matter: evolutionary forces driving nuclear genome miniaturization and expansion. Ann Bot. 95:147–175.
Curtiss, CC., Langlois, GW., Busse, LB., Mazzillo, F. & Silver, MW. 2008. The emergence of Cochlodinium along the California Coast (USA). Harmful Algae. 7:337–346.
Dolezel, J., Bartos, J., Voglmayr, H. & Greilhuber, J. 2003. Nuclear DNA content and genome size of trout and human. Cytometry A. 51:127–128.
Dorantes-Aranda, JJ., García-de la Parra, LM., Alonso-Rodríguez, R., Morquecho, L. & Voltolina, D. 2010. Toxic effect of the harmful dinoflagellate Cochlodinium polykrikoides on the spotted rose snapper Lutjanus guttatus. Environ Toxicol. 25:319–326.
Ebenezer, V. & Ki, J-S. 2014. Biocide sodium hypochlorite decreases pigment production and induces oxidative damage in the harmful dinoflagellate Cochlodinium polykrikoides. Algae. 29:311–319.
Elliott, TA. & Gregory, TR. 2015. What’s in a genome? The C-value enigma and the evolution of eukaryotic genome content. Philos Trans R Soc Lond B Biol Sci. 370:20140331 pp.
Gárate-Lizárraga, I. 2013. Bloom of Cochlodinium polykrikoides (Dinophyceae: Gymnodiniales) in Bahía de La Paz, Gulf of California. Mar Pollut Bull. 67:217–222.
Gobler, CJ., Berry, DL., Anderson, OR., Burson, A., Koch, F., Rodgers, BS., Moore, LK., Goleski, JA., Allam, B., Bowser, P., Tang, Y. & Nuzzi, R. 2008. Characterization, dynamics, and ecological impacts of harmful Cochlodinium polykrikoides blooms on eastern Long Island, NY, USA. Harmful Algae. 7:293–307.
Gregory, SG., Sekhon, M., Schein, J., Zhao, S., Osoegawa, K., Scott, CE., Evans, RS., Burridge, PW., Cox, TV., Fox, CA., Hutton, RD., Mullenger, IR., Phillips, KJ., Smith, J., Stalker, J., Threadgold, GJ., Birney, E., Wylie, K., Chinwalla, A., Wallis, J., Hillier, L., Carter, J., Gaige, T., Jaeger, S., Kremitzki, C., Layman, D., Maas, J., McGrane, R., Mead, K., Walker, R., Jones, S., Smith, M., Asano, J., Bosdet, I., Chan, S., Chittaranjan, S., Chiu, R., Fjell, C., Fuhrmann, D., Girn, N., Gray, C., Guin, R., Hsiao, L., Krzywinski, M., Kutsche, R., Lee, SS., Mathewson, C., McLeavy, C., Messervier, S., Ness, S., Pandoh, P., Prabhu, A-L., Saeedi, P., Smailus, D., Spence, L., Stott, J., Taylor, S., Terpstra, W., Tsai, M., Vardy, J., Wye, N., Yang, G., Shatsman, S., Ayodeji, B., Geer, K., Tsegaye, G., Shvartsbeyn, A., Gebregeorgis, E., Krol, M., Russell, D., Overton, L., Malek, JA., Holmes, M., Heaney, M., Shetty, J., Feldblyum, T., Nierman, WC., Catanese, JJ., Hubbard, T., Waterston, RH., Rogers, J., de Jong, PJ., Fraser, CM., Marra, M., McPherson, JD. & Bentley, DR. 2002. A physical map of the mouse genome. Nature. 418:743–750.
Gregory, TR. 2001. Coincidence, coevolution, or causation? DNA content, cell size, and the C-value enigma. Biol Rev Camb Philos Soc. 76:65–101.
Gregory, TR. 2002. A bird’s-eye view of the C-value enigma: genome size, cell size, and metabolic rate in the class aves. Evolution. 56:121–130.
Gregory, TR., Nicol, JA., Tamm, H., Kullman, B., Kullman, K., Leitch, IJ., Murray, BG., Kapraun, DF., Greilhuber, J. & Bennett, MD. 2007. Eukaryotic genome size databases. Nucleic Acids Res. 35(Suppl 1):D332–D338.
Gregory, TR. & Witt, JDS. 2008. Population size and genome size in fishes: a closer look. Genome. 51:309–313.
Guillard, RRL. & Ryther, JH. 1962. Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt, and Detonula confervacea (Cleve) Gran. Can J Microbiol. 8:229–239.
Guo, R., Wang, H., Suh, YS. & Ki, J-S. 2016. Transcriptomic profiles reveal the genome-wide responses of the harmful dinoflagellate Cochlodinium polykrikoides when exposed to the algicide copper sulfate. BMC Genomics. 17:29 pp.
Hare, EE. & Johnston, JS. 2011. Genome size determination using flow cytometry of propidium iodide-stained nuclei. In : Hare EE, Johnston JS, editors Molecular Methods for Evolutionary Genetics. 772:Humana Press, NY, 3–12.
Kang, EJ., Kim, J-H., Kim, K., Choi, H-G. & Kim, KY. 2014. Re-evaluation of green tide-forming species in the Yellow Sea. Algae. 29:267–277.
LaJeunesse, TC., Lambert, G., Andersen, RA., Coffroth, MA. & Galbraith, DW. 2005. Symbiodinium (Pyrrhophyta) genome sizes (DNA content) are smallest among dinoflagellates. J Phycol. 41:880–886.
Lin, S., Cheng, S., Song, B., Zhong, X., Lin, X., Li, W., Li, L., Zhang, Y., Zhang, H., Ji, Z., Cai, M., Zhuang, Y., Shi, X., Lin, L., Wang, L., Wang, Z., Liu, X., Yu, S., Zeng, P., Hao, H., Zou, Q., Chen, C., Li, Y., Wang, Y., Xu, C., Meng, S., Xu, X., Wang, J., Yang, H., Campbell, DA., Sturm, NR., Dagenais-Bellefeuille, S. & Morse, D. 2015. The Symbiodinium kawagutii genome illuminates dinoflagellate gene expression and coral symbiosis. Science. 350:691–694.
Richlen, ML., Morton, SL., Jamali, EA., Rajan, A. & Anderson, DM. 2010. The catastrophic 2008–2009 red tide in the Arabian gulf region, with observations on the identification and phylogeny of the fish-killing dinoflagellate Cochlodinium polykrikoides. Harmful Algae. 9:163–172.
Shoguchi, E., Shinzato, C., Kawashima, T., Gyoja, F., Mungpakdee, S., Koyanagi, R., Takeuchi, T., Hisata, K., Tanaka, M., Fujiwara, M., Hamada, M., Seidi, A., Fujie, M., Usami, T., Goto, H., Yamasaki, S., Arakaki, N., Suzuki, Y., Sugano, S., Toyoda, A., Kuroki, Y., Fujiyama, A., Medina, M., Coffroth, MA., Bhattacharya, D. & Satoh, N. 2013. Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr Biol. 23:1399–1408.
Thornhill, DJ., Lajeunesse, TC. & Santos, SR. 2007. Measuring rDNA diversity in eukaryotic microbial systems: how intragenomic variation, pseudogenes, and PCR artifacts confound biodiversity estimates. Mol Ecol. 16:5326–5340.
Veldhuis, MJW., Cucci, TL. & Sieracki, ME. 1997. Cellular DNA content of marine phytoplankton using two new fluorochromes: taxonomic and ecological implications. J Phycol. 33:527–541.
Vinogradov, AE. 2004. Evolution of genome size: multilevel selection, mutation bias or dynamical chaos? Curr Opin Genet Dev. 14:620–626.
Wisecaver, JH. & Hackett, JD. 2011. Dinoflagellate genome evolution. Annu Rev Microbiol. 65:369–387.
|
|
|||||||||||||||||||||||||||||||||||||||