INTRODUCTION
Genetic studies are increasingly exploring the patterns of diversity, evolutionary history, and phylogeography for many marine species (Bowen et al. 2016, Faria et al. 2021). Phylogeographic analyses are often used to trace the process of marine species dispersal and population connectivity (e.g., Zuccarello and Martin 2016, Vieira et al. 2021), but can also detect cryptic species diversity and biogeographic breaks in the ocean (Fraser et al. 2013, Muangmai et al. 2015a). Such analyses are crucial to provide a useful framework for marine natural bioresource management and invasive species risk assessment (Teske et al. 2011, Bowen et al. 2016).
In the Southeast Asia region, genetic diversity and phylogeographic patterns of macroalgae are being increasingly documented. Many previous studies indicated different levels of genetic diversity among macroalgal species. For example, the brown alga Sargassum polycystum C. Agardh showed low genetic diversity, but higher diversity on the Andaman coast of Thailand (Kantachumpoo et al. 2014), whereas, in the Philippines, high genetic diversity was detected in the red algae, Phycocalidia acanthophora (E. C. Oliveira & Coll) Santiañez (Dumilag and Aguinaldo 2017, as Pyropia acanthophora) and Gracilaria salicornia (C. Agardh) E. Y. Dawson (Ferrer et al. 2019). Phylogeographic studies also indicated partial isolation between the Indian and Pacific Ocean populations of S. polycystum (Kantachumpoo et al. 2014). This separation appears to be maintained by a geographic barrier (the Thai-Malay Peninsula) and the current oceanographic discontinuity between the Indian and Pacific oceans (Kantachumpoo et al. 2014, Wichachucherd et al. 2014).
Cryptic species are common in red algae (e.g., Zuccarello et al. 2002, Muangmai et al. 2014, Díaz-Tapia et al. 2018) and are known to differ in many species-specific properties, although being morphologically identical. Their demographic histories can differ (Muangmai et al. 2015a, 2022) and they can differ physiologically (Muangmai et al. 2015b) and chemically (Payo et al. 2011, Bracegirdle et al. 2019). In addition, they can differ in their intertidal position (Muangmai et al. 2016). In the Indo-Pacific, genetic studies revealed cryptic diversity in many red seaweeds (e.g., Payo et al. 2013, Gabriel et al. 2016, Saengkaew et al. 2016). Recognizing this cryptic algal diversity highlights a substantial undiscovered biodiversity, with implication for a need to continue a thorough investigation of red algae using data based on extensive markers and sampling.
Bostrychia tenella (J. V. Lamouroux) J. Agardh is a marine filamentous red alga, which is distributed in tropical and subtropical regions (King and Puttock 1989, Zuccarello et al. 2015). This species naturally grows either on mangroves or in the high intertidal on rocks. Zuccarello et al. (2015) were able to resurrect the species B. binderi Harvey from B. tenella using DNA-based species delimitation, but also showed cryptic species diversity within B. tenella, consisting of two species B and C with overlapping distributions. In Thailand, B. tenella is relatively common and widespread across the coastal areas based on morphological record (Lewmanomont et al. 1995). Recent genetic analysis indicated the occurrence of B. tenella species B and C on both eastern (the Gulf of Thailand – Pacific Ocean) and western coasts (Andaman Sea – Indian Ocean) of Thailand (Saengkaew et al. 2016). Additionally, coexistence of two cryptic species was reported from both coastal regions of the country (Saengkaew et al. 2016). However, due to the limited sampling this study mainly focused on the phylogenetic diversity of B. tenella, and therefore genetic diversity and distribution pattern of this cryptic species across the Thai-Malay Peninsula have not been fully examined.
In this present study, we used the chloroplast-encoded RuBisCo spacer of B. tenellla across the Thai-Malay Peninsula to assess its genetic diversity and phylogeographic pattern. We also wanted to see if there were genetic differences of these cryptic species between the Andaman coasts and the Gulf of Thailand, and then determined what factors could explain these distribution patterns.
MATERIALS AND METHODS
Algal sampling
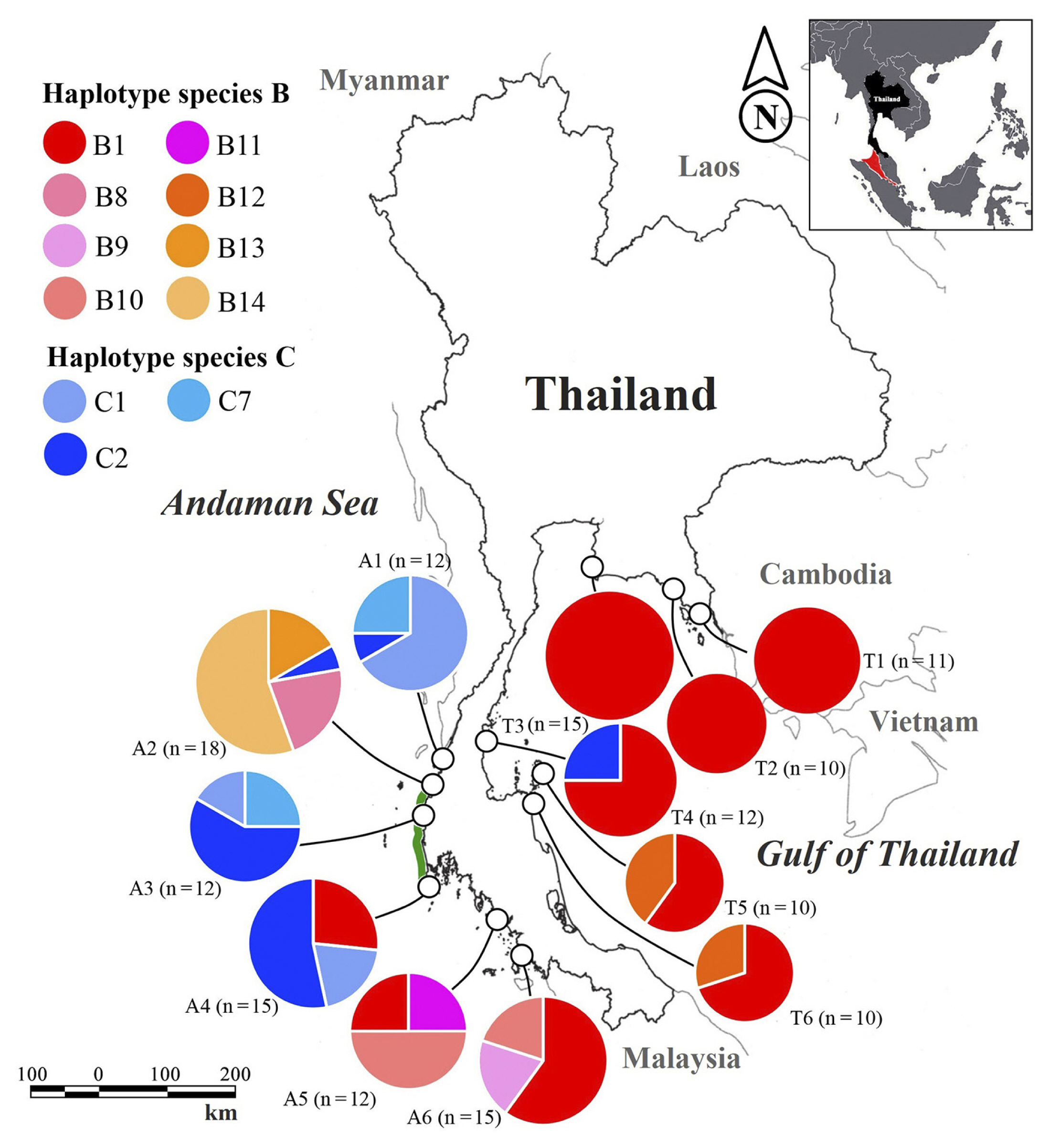
We surveyed B. tenella populations around rocky intertidal areas of 12 sites along the Thai-Malay Peninsula: the Gulf of Thailand (T1–T6) and the Andaman Sea (A1–A6) (Table 1, Fig. 1), between 2017 and 2019. At each site, algal samples were randomly collected, at least 10 samples per population, and preserved in silica gel for both morphological and molecular analyses. For morphological observation, preserved samples were prepared by soaking in water for 5 min and subsequently simultaneously stained and preserved in 1% aniline blue acidified with 1% HCl and mounted in 50% glucose syrup (Karo Corn Syrup) on microscope slides. Species identification was based on previous publications (King and Puttock 1989, Zuccarello et al. 2015).
DNA extraction, polymerase chain reaction, and sequencing
Genomic DNA isolation was performed using apical portions of dried algal specimens, with a modified Chelex extraction method (Zuccarello et al. 1999) or Norgen Biotek’s Plant/Fungi DNA Isolation Kit (Norgen Biotek Corp., Thorold, Canada). The plastid-encoded RuBisCo spacer was selected for polymerase chain reaction (PCR) amplification. This fragment is a powerful region for exploring inter- and intra-species diversity in Bostrychia (Zuccarello and West 2003, Zuccarello et al. 2015). PCR was carried out using PCR Master mix Solution (i-Taq, iNtRON Biotechnology DR, Seongnam, Korea), in a total volume of 20 μL, consisting of 10 μL of i-Taq, 10 pmol of each primer and 2 μL of genomic DNA (~10–20 ng). PCR amplification profile and procedure followed Zuccarello and West (2003). All amplified products were cleaned using ExoSAP-IT (USB, Cleveland, OH, USA) and then sequenced commercially in both directions using PCR primers (U2Bio Inc., Seoul, Korea).
Alignment, phylogenetic reconstructions, historical demography, and population structure analysis
Sequences were edited, assembled, and aligned using the Geneious Prime software package (Biomatters, available from http://www.geneious.com/). Alignments of the RuBisCo spacer sequences were performed using the MAFFT algorithm (Katoh et al. 2002) and were further manually refined. Additional 12 sequences of B. tenella were obtained from GenBank and included in the alignment (Supplementary Table S1).
Phylogenetic trees were reconstructed using maximum-likelihood (ML) and Bayesian inference (BI) methods using IQ-TREE (Minh et al. 2020) and MrBayes v3.2 (Ronquist et al. 2012), respectively. ML analyses were carried out with the HKY + G model as selected by ModelFinder (Kalyaanamoorthy et al. 2017) and 1,000 bootstrap replicates. The molecular evolution models for BI were selected using Kakusan 4 (Tanabe 2011). BI analyses were performed using a GTR + I + R model, with two parallel runs of four Markov chains for a million generations. We sampled one tree every 1,000 generations and then removed 2,500 trees (burn-in) before determining a consensus tree and posterior probabilities. Both ML and BI trees were edited with the program FigTree v1.4.4 (Rambaut 2016).
Number of haplotypes (H), haplotype (Hd), and nucleotide (π) diversity were calculated for each population using DnaSP v6 (Rozas et al. 2017). Haplotype networks were produced using median joining generated in PopART v1.7 (University of Otago, available from http://popart.otago.ac.nz.). Additionally, the historical demography for each species (species B vs. species C) and population (Andaman Sea vs. the Gulf of Thailand) levels was estimated by the analyses of mismatch distribution and neutrality tests, Tajima’s D (Tajima 1989) and Fu’s FS, as implemented in DnaSP. The analysis of mismatch distribution uncovers the demographic history of populations, with population expansions producing a unimodal distribution, whereas stable population show a multimodal distribution (Rogers and Harpending 1992).
Population differentiation was analyzed separately for each cryptic species for populations with at least 10 samples. The analysis was examined using two different approaches. First, pairwise population differentiation (FST) values were estimated with a significance level of 0.05 determined by 10,000 permutations. Secondly, Analysis of Molecular Variance (AMOVA) was performed with significance determined by 1,023 permutations. All analyses were conducted in Arlequin v 3.5.1.3 (Excoffier and Lischer 2010).
RESULTS
Genetic diversity and distribution
The RuBisCo spacer with partial 3′-rbcL and 5′-rbcS gene sequences of 275–286 bp were successfully generated from 152 samples of 12 populations of B. tenella from both coasts of Thailand. Genetic distance among these sequences ranged from 0.0 to 6.6%. ML and BI analyses yielded almost complete topological congruence, and the ML tree is presented in Supplementary Fig. S1. Phylogenetic analyses demonstrated the occurrence of two different cryptic species of B. tenella: B and C, based on Zuccarello et al. (2015), in Thailand. Among 152 samples, 113 samples were species B, while 39 samples belonged to species C. Intraspecific genetic variation varied from 0.0–3.4% for species B and ranged from 0.0–1.3% for species C.
Haplotype and genetic diversity indices of the two cryptic species are presented in Table 1. Within species, both cryptic B. tenella species revealed a moderate genetic diversity (Hd = 0.670 for species B and Hd = 0.629 for species C), but higher nucleotide diversity for species B (π = 0.00807) than for species C (π = 0.00272) (Table 1). Within populations haplotype diversity (Hd) ranged from 0.00 to 0.68 for species B and from 0.00 to 0.62 for species C (Table 1). Nucleotide diversity (π) was relatively low, varying from 0.0000 to 0.0037 for species B, and from 0.0000 to 0.0023 for species C (Table 1).
Both cryptic B. tenella species were found on both coastal areas of Thailand. Cryptic species B was widely distributed in Thailand, recorded in 9 out of 12 sampled populations, while species C was detected in 7 populations (Fig. 1). Additionally, both cryptic species were found to coexist at three of the 12 sites (A2 and A4 in the Andaman Sea and T4 from the Gulf of Thailand) (Fig. 1).
Haplotype diversity and networks
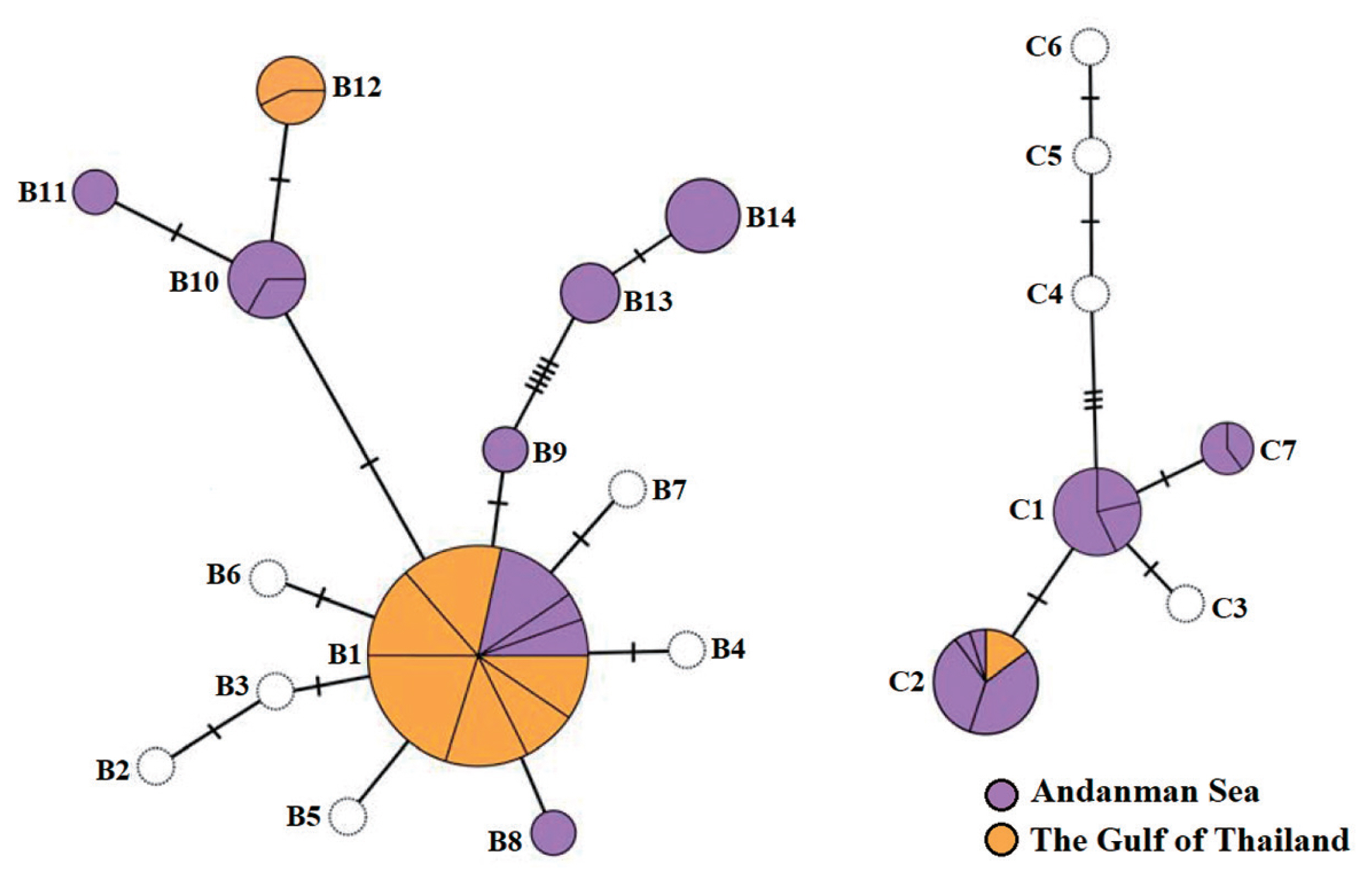
Median-joining haplotype networks of cryptic species B and C constructed using RuBisCo spacer sequence (including haplotypes from Zuccarello et al. 2015 and Saengkaew et al. 2016) are presented in Fig. 2. Cryptic species B consisted of eight haplotypes in Thailand (B1, B8–B14) of which the B9–B14 are reported here for the first time. The haplotype network of species B was star-like with a central common haplotype B1 (80% of samples), found in nine of 12 populations sampled in the present study (Table 1, Fig. 1) and in addition at three sampled sites (Phuket, Chumphon and Nakhon Si Thammarat) of Saengkeaw et al. (2016) and in Sabah, Malaysia (Zuccarello et al. 2015). Five novel B-haplotypes were only found in Andaman populations (A2, A5, and A6), while haplotype B12 was restricted to the Gulf of Thailand (populations T5 and T6) (Table 1, Fig. 1).
Cryptic species C is represented by three haplotypes in Thailand (C1, C2, and C7) (Fig. 2). Haplotype C2 was relatively common, accounting for 51% of all samples, and it occurred in all five populations where this species was found. C2 is the only haplotype of cryptic species C which was shared on both coasts of Thailand, while the other haplotypes were confined to the Andaman Sea (Table 1, Fig. 1). Haplotype C1 was shared in nearly all population of this species in the Andaman Sea, while haplotype C7 was detected from populations A1 and A3 (Table 1, Fig. 1).
Demographic history and population differentiation
The mismatch distribution showed a multimodal pairwise distance distribution of species B, but unimodal distribution of species C, indicating that species C has undergone a recent expansion (Supplementary Fig. S2). These demographic patterns were further supported by significant positive values of Tajima’s D test for species B (0.08932, p < 0.05), suggesting a stable population, but significant negative value for species C (−1.11856, p < 0.05), indicating a population expansion. However, the estimates of Fu’s FS were not significant (1.12591, p > 0.1 for species B, and 1.02017, p > 0.1 for species C).
Analyses within biogeographic regions were only done for B. tenella species B. Species B from the Andaman Sea displayed a multimodal distribution (Supplementary Fig. S2) and significant positive value of Fu’s FS test (1.78007, p < 0.05), whereas species B from the Gulf of Thailand showed a unimodal distribution (Supplementary Fig. S2) plus negative values of Fu’s FS test (−0.54417, p > 0.1), implying that the Gulf populations has more likely undergone recent expansion than the Andaman Sea populations. Values of Tajima’s D test were not significant (1.74664, p > 0.1 for Andaman Sea population and 1.34228, p > 0.1 for the Gulf population).
Genetic differentiation (FST values) was moderate to low between populations of B. tenella species B from the Andaman Sea (A2, A5, and A6) and the Gulf of Thailand (T1–T6), with pairwise FST values ranging between 0.14 and 0.69 (p < 0.05) (Supplementary Table S2). Significant moderate to low genetic differentiation was detected within Andaman Sea populations, with FST value in the range of 0.67 of 0.28 (A2, A5, and A6). While low genetic differentiation was discovered from the Gulf of Thailand population, with FST value in the range of 0.08 to 0.40 (Supplementary Table S2). No genetic differentiation (FST = 0.00) was found among the upper Gulf populations (T1–T4), while a slight genetic differentiation (FST = 0.08) was observed between the lower Gulf populations (T5 and T6) (Supplementary Table S2).
Pairwise FST values indicated that Andaman Sea populations of cryptic species C differed significantly. Population A1 was genetically distinct from the other two populations, with FST values ranging between 0.40 to 0.51. Low genetic differentiation was observed between populations A3 and A4 (FST = 0.03) (Supplementary Table S3).
The AMOVA was calculated to determine genetic differentiation of cryptic species B between the Andaman Sea (A2, A5, and A6) and the Gulf of Thailand (T1–T6) populations. AMOVA results indicated that the proportion of variation attributed to among populations within regions was 47.70% and within-population differences for 32.43%, whereas 19.87% occurred among regions (Supplementary Table S4). The F statistics revealed the significant and high genetic differences within populations (FST = 0.67), but low among regions (FCT = 0.19) (Supplementary Table S4).
DISCUSSION
With wide-range sampling along the coasts of Thailand and chloroplast DNA analysis, we reconfirmed the occurrence of two distinct lineages, B and C, of B. tenella on both coastal regions of the country. In addition, we detected differences between these two cryptic species. Bostrychia tenella species B showed higher abundance and genetic diversity than species C. Different levels of genetic diversity between cryptic species was also observed in other red seaweeds, e.g., Bostrychia intricata (Bory) Montagne (Muangmai et al. 2015a, 2022), Sarcopeltis skottsbergii (Setchell & N. L. Gardner) Hommersand, Hughey, Leister & P. W. Gabrielson (Billard et al. 2015, as Gigartina skottsbergii), and Asparagopsis taxiformis (Delile) Trevisan (Zanolla et al. 2018). These previous studies suggested that these different levels of genetic variation among cryptic species was likely due to different evolutionary and demographic histories. Our additional analyses of mismatch distribution and Tajima’s D test further supported the different patterns of historic population demography in these two cryptic species of B. tenella. We, therefore, postulate that cryptic B. tenella species B and C in this area have different patterns of historical population demography. However, the evolutionary history of cryptic B. tenella species needs further investigation with larger sample sizes and more variable markers (e.g., microsatellites).
We also observed different levels of genetic diversity in cryptic B. tenella species between the Andaman Sea and the Gulf of Thailand. Our data showed that genetic diversity of Andaman Sea populations of B. tenella was higher than that of the populations in the Gulf of Thailand. Additionally, we identified several novel haplotypes of cryptic species B from the Andaman Sea, whereas only one new haplotype was found in the Gulf of Thailand. This is consistent with previous findings that the Andaman Sea harbored greater diversity, both genetic and at the species levels than the Gulf of Thailand for seaweed (e.g., Kantachumpoo et al. 2014, Wichachucherd et al. 2014, Pongparadon et al. 2015) and other marine species, for example littoral earthworms (Seesamut et al. 2019), dugongs (Poommouang et al. 2021) and clams (Suppapan et al. 2021). Differences in genetic diversity across the Thai-Malay Peninsula could be driven by historical environmental changes and different geographical features. The lowering of sea levels (~120 m below present-day sea level) during the Last Glacial Period (ca. 17,000 years ago) resulted in the formation of landmass in Southeast Asia known as Sundaland (Sathiamurthy and Voris 2006). The emergent Sundaland would have eliminated the habitat availability of marine taxa through the Gulf of Thailand and constrained populations to the east (South China Sea) and west (the Andaman Sea) coasts of the landmass (Cannon et al. 2009, Ludt and Rocha 2015). By the end of the last glaciation (ca. 11,000 years ago), recolonization of the Gulf of Thailand would have led to a potential founder event for these populations (Palmer 2004, Guo et al. 2020). Accordingly, this would result in higher genetic diversity in Andaman Sea populations, and lower diversity in the Gulf population. Such a hypothesis has been applied to mangrove species in Southeast Asia (see Wee et al. 2014, Guo et al. 2020), and therefore could also apply to B. tenella and other seaweed around the Thai-Malay Peninsula.
Although previous studies have revealed strong phylogeographic patterns between the Gulf of Thailand and Andaman Sea in some marine species (e.g., Nguyen et al. 2014, Wichachucherd et al. 2014, Seesamut et al. 2019, Panithanarak 2020), we did not find a clear phylogeographic signal in the B. tenella species complex across the Thai-Malay Peninsula. Our results showed that pairwise FST value between the Andaman Sea and the Gulf populations of cryptic species B demonstrated the presences of low to moderate levels of genetic differentiation, with only 19.87% of detected genetic differences found between the Andaman Sea and the Gulf of Thailand (AMOVA results), indicating moderate to high connectivity in cryptic species B. Our findings suggest the occurrence of gene flow around the Thai-Malay Peninsula. Furthermore, our demographic history analyses indicated that the Gulf populations of cryptic species B historically experienced a more recent expansion than Andaman Sea populations. Accordingly, we assume that the connectivity of B. tenella species B population across Thai-Malay Peninsula could possibly be associated with the historical subsidence of Sundaland, leading to a recent population expansion of this alga during the inundation of the Gulf of Thailand, and regional circulation patterns linking these two coastal areas (Haditiar et al. 2020).
In the Andaman Sea, we detected some genetic differences between B. tenella species. Cryptic species B population from Ranong province (A2) differed significantly from Trang and Satun provinces (A5 and A6). Additionally, analysis of haplotype distribution indicated that haplotype B1 found in population A4 was also found in A5 and A6, but not in A2, implying the genetic break occurring north to Phuket provinces (A4) (Fig. 1). Similarly, pairwise FST showed moderate genetic differentiation of cryptic species C populations between Ranong province (A1) and Phang Nga and Phuket provinces (A3 and A4). These results indicate that there is low gene flow between these populations of both cryptic species. Potential factors contributing to the genetic differentiation could be attributed to the ocean circulation patterns from North Indian Ocean and the Malacca Strait (Wyrtki 1961, Chatterjee et al. 2017). The North Indian Ocean current always flows southwesterly toward the Andaman Sea, while the Malacca Strait current directs north-westward toward the Andaman Sea. These two currents flowed offshore and partly mix during both the North–East (December through February) and the South–West (June through September) monsoon season (Rizal et al. 2012). This current pattern may act as a marine barrier to gene flow, which promotes isolation between Andaman Sea populations. Such potential barrier was speculated to contribute to restricted distribution of some Halimeda species (Pongparadon et al. 2015) and genetic divergence of sandfish (Ninwichian and Klinbunga 2020) and mangrove (Guo et al. 2020).
Our results also showed the coexistence of cryptic B. tenella species B and C in populations of the Andaman Sea and the Gulf of Thailand. Coexistence of different cryptic species is likely to be common in the genus Bostrychia (Zuccarello and West 2003, 2011, Muangmai et al. 2014, 2022). How different cryptic Bostrychia species can coexist could possibly be explained by their eco-physiological differences, including difference in tidal height distribution, wave exposure and salinity tolerance (Muangmai et al. 2015b, 2016). In order to obtain detailed insight into the forces maintaining the coexistence in cryptic B. tenella species, their ecological interactions and physiological aspects need further investigation.
In conclusion, our data clearly indicated the contrasting patterns of genetic diversity between two cryptic B. tenella species B and C in Thai waters, suggesting different demographic history between these two cryptic species. However, both cryptic species from Andaman Sea possessed higher level of genetic diversity than the Gulf of Thailand populations. These findings have implications for not only evaluating the potential genetic resources of these cryptic red algae, but even contributing to accurate marine biodiversity assessment for strategic conservation planning of coastal areas. We also detected weak phylogeographic structure of B. tenella species B between the two coasts, but a relatively strong phylogeographic patterning of B. tenella species C along the Andaman Sea coast. These patterns could have been shaped by a combination of historical geological events (submergence of Sundaland) and contemporary regional circulation. However, in order to better understand the phylogeographic structure and demographic history of cryptic B. tenella species distributed around the Thai-Malay Peninsula, additional sampling sites from the Malacca Strait and the South China Sea, plus applying more variable genetic markers are necessary.