ABSTRACTCryptic stages of diverse macroalgae present in natural substrata, “the bank of microscopic forms”, were isolated into clonal cultures and identified based on both morphological characteristics and DNA barcoding. Approximately 120 clonal isolates from 308 natural substratum samples were collected from the entire coastline of Kuwait. Amongst these isolates, 77 (64%) were identified through DNA barcoding using the nuclear ribosomal small subunit, RuBisCO spacer (ITS2, tufa, rbcL, psaA, and psbA) and sequencing. Twenty-six isolates (34%) were identified in the division Chlorophyta, 18 (23%) as Phaeophyceae, and 33 (43%) as Rhodophyta. For all DNA sequences in this study, species-level cut off applied was ≥98% homology which depend entirely on the markers used. Three putative new records of Chlorophyta new for the Arabian Gulf were made: Cladophora laetevirens (Dillwyn) Kützing, Ulva torta (Mertens) Trevisan and Ulvella leptochaete (Huber) R. Nielsen, C. J. O′Kelly & B. Wysor in Nielsen, while Cladophora gracilis Kützing and Ulva ohnoi M. Hiraoka & S. Shimada are new records for Kuwait. For Phaeophyceae, Ectocarpus subulatus Kützing and Elachista stellaris Areschoug were new records for the Gulf and Kuwait. In the Rhodophyta, Acrochaetium secundatum (Lyngbye) Nägeli in Nägeli & Cramer, Ceramium affine Setchell & N. L. Gardner, Gelidium pusillum var. pakistanicum Afaq-Husain & Shameel and Dasya caraibica Børgesen are new records for the Gulf and Kuwait, while the red alga Stylonema alsidii (Zanardini) K. Drew is a new record for Kuwait. Several isolates identified corresponded to genera not previously reported in Kuwait and / or the Arabian Gulf, such as Porphyrostromium Trevisan, a new genus from the Bangiales, and two unidentified species for the Planophilaceae Škaloud & Leliaert. The isolates cultivated from substrata enhance understanding of the marine macroalgal diversity in the region and confirmed that the Germling Emergence Method is suitable for determining the actual diversity of a given study area through isolation from cryptic life-history phases.
INTRODUCTIONGlobally, numerous shorelines are yet to be explored for benthic marine algal diversity (Wynne et al. 2020). Marine algae are important shelter or habitats for sessile - micro- and macro-organisms and are the basis of food webs in marine ecosystems (Wolf 2012, Macreadie et al. 2017). However, only approximately 10% of algal diversity have been discovered and formally described, with a lack of knowledge especially in remote regions (De Vargas et al. 2015, Bartolo et al. 2020). Seaweeds produce reproductive cells or multicellular propagules at maturity, many of which will attach to available substrata and, after a period of time, form a “bank of algal microscopic forms” (Hoffmann and Santelices 1991, Peters et al. 2015, Schoenrock et al. 2021). However, these cryptic life forms are difficult to capture and characterize.
In the last 20–30 years, the development of in vitro culture techniques for isolates from incubated natural substrata, such as small pebbles and sand grains collected during fieldwork or diving surveys, has helped in the investigation of macroalgal microscopic stages from remote and under sampled areas (Ramírez and Müller 1991, Müller and Ramírez 1994, Peters et al. 2015). The Germling Emergence (GE) technique has the potential to discover cryptic species and new records of macroalgal taxa by a combination of morphological characterization with DNA barcoding. Comparing short DNA barcodes with available reference sequences in public databases can lead to accurate identification of cultured microscopic species or microstages that can be difficult to classify morphologically (Peters et al. 2015, Bartolo et al. 2020). For example, Zuccarello et al. (2011) and West et al. (2012) reviewed the order Erythropeltidales (Rhodophyta) using a combination of the GE method and molecular data that enabled new records and discoveries of macroalgal taxa around the world. These discoveries included, for example, a novel Desmarestia species and a number of Ectocarpales species from the Canadian Arctic (Küpper et al. 2016), two novel members of the Pelagophyceae, Sungminbooa kuepperi R. A. Andersen & B. Melkonian from the Beagle Channel, Tierra del Fuego, and Chrysoreinhardia muelleri M. Melkonian & R. A. Andersen from the Falklands (Han et al. 2018). Peters et al. (2015) used the GE method combined with 5′-cytochrome oxidase (COI) barcoding to detect cryptic diversity (including three putative new species) within Ectocarpales collected from different geographical regions. Additionally, in circalittoral waters (24 m depth) of the Mediterranean Sea, the GE method succeeded in isolating the minute benthic multicellular alga Schizocladia ischiensis; thus, this method appears to reveal benthic algae even when small in size (Rizouli et al. 2020). In another study, isolates of the genus Ectocarpus and associated bacterial strains from Hopkins River in Australia, obtained using the GE method, provided the possibility to study Ectocarpus adaptations to abiotic parameters - especially low salinity and the interactions of the alga with the endogenous microbiome (Dittami et al. 2020b). Recently, the GE method enabled the description of a new species of red alga from the Celebes Sea, Hypoglossum sabahense (Wynne et al. 2020).
The aim of the work presented here was to identify algal isolates from a collection of microscopic forms from different locations in Kuwait, obtained using the GE technique. A few additional algal cultures were obtained from fragments or reproductive cells of macroscopic algae. Molecular identification and establishment of gene phylogenies were carried out using commonly employed markers, for which there were sufficient reference sequences in public databases. The mitochondrial COI is a useful barcode marker since it discriminates well between species and it has become a preferred locus in DNA barcoding of brown and red macroalgae (Saunders and McDevit 2013). The plastid-encoded large and small subunits of ribulose-1,5-bisphosphate carboxylase (rbcL and rbcS, respectively) are separated by a spacer, which is variable in length (Destombe and Douglas 1991). rbcL is also among the preferred loci for phylogenetic studies and DNA barcoding of brown macroalgae (Siemer et al. 1998, Saunders and McDevit 2013), although it is slightly less discriminatory than COI between species of Phaeophyceae. The RuBisCO spacer is highly variable and cannot serve for phylogenetic analyses because of limited alignability across species and genera. Sequences of the spacer, however, are occasionally published together with rbcL sequences and can be used as barcodes for the identification of species (Siemer et al. 1998).
MATERIALS AND METHODSField sites, specimens, and laboratory cultureSamples were collected during late winter and spring (February and April 2019), from 38 sampling points (sites) in 12 localities representing the different maritime regions from Northern and Southern Provinces in Kuwait including offshore islands (Fig. 1). Of the 308 total individual samples, 261 (85%) were abiotic (small fragments of rock, or pebbles) while 47 (15%) were biotic substrata (fragments of subtidal macroflora, such as seagrass leaves and Sargassum blades). Generally, samples were collected from each sector at 3 transects; upper (closest to the shore) middle, lower intertidal zone and rocky shore at each location during low tide. Furthermore, substratum samples were collected from several of the islands of Kuwait by SCUBA diving in the shallow subtidal zone around Failaka and Umm al-Maradim (15 m), Qaruh (6 m), and Kubbar (10–35 m) (Supplementary Table S1). Also, the sampling were covered various distances from brine outfalls at typical two desalination plants, namely; Doha East (DE) and Al-Zour South (ZS). Triplicate samples of natural substrata were placed in 15 mL FALCON tubes previously filled with 10 mL heat-sterilized sea water, before transport to the Bezhin Rosko Laboratory of AFP (Santec, France). There, the contents of each tube were transferred to 55 mm diameter Petri dishes containing 8 mL Provasoli-enriched natural autoclaved sea water culture medium (Provasoli 1966, Starr and Zeikus 1993, Tarakhovskaya et al. 2012) and incubated at 15°C in natural daylight at a north-facing window for 2–6 weeks. In order to prevent diatom growth, germanium dioxide (GeO2, 3–4 mg L−1) was added to the medium (Peters et al. 2015). After three to four weeks, unialgal isolates were obtained from the dishes by cutting accessible parts of germlings with a sharp, freshly snapped, glass Pasteur pipet tip and transferred into fresh culture medium without GeO2 (Lewin 1966). The laboratory culture followed standard methods (Coelho et al. 2012), with monthly serial transfer. The temperature was maintained at a constant 25 ± 1°C in a CO2 incubator (240 L MIR-253-CFC FREE; Sanyo, UK), irradiance was by dimmed natural light (less than 40 μmol m−2 s−1 photon fluence rate) measured with a Skye Instruments SKP 200 light meter (Skye Instruments Ltd., Llandrindod Wells, Wales, UK) and daylengths were based on the natural change. To minimize duplicate isolation of the same species, multiple isolates were made from each dish only when algae exhibited different morphologies. Strains that were not single cultures after a further two months of cultivation were excluded from subsequent analyses. The February and April isolates were deposited in collections at the University of Aberdeen, Scotland, UK, in the Bezhin Rosko Laboratory (France) and in the Algal Culture Unit (KUAC) at Kuwait University, Kuwait.
Molecular work and phylogenetic analysesDNA was extracted from 5–15 mg wet weight of algal cultures using CTAB, and the GENEJET Plant Genomic DNA Purification Kit (Thermo Scientific, Vilnius, Lithuania) according to the manufacturer’s protocol (Gachon et al. 2009). Extracted DNA from the unialgal isolates was amplified by polymerase chain reaction (PCR) and sequenced. First, the nuclear ribosomal small subunit nrSSU locus was used to confirm that the extracted DNA was suitable for amplification. However, this marker did not discriminate sufficiently between species, and barcode markers with higher resolution were used for final identification (Bartolo et al. 2020). Therefore, several markers were used together with the nrSSU locus to obtain species-level identification. Internal transcribed spacer 1 (ITS1), internal transcribed spacer 2 (ITS2), and plastid elongation factor (tufA) locus used for Chlorophyta, the partial mitochondrial gene regions (5’COI) to examine Phaeophyceae and plastid-encoded psaA, psbA gene and plastid locus, such as RuBisCO spacer region rbcL were used for Rhodophyta (Table 1). Two markers were used for isolates of Chlorophyta, the plastid-encoded DNA barcode marker elongation factor tufa and the nuclear ITS2. tufA has been used to discriminate amongst green algal species (Kirkendale et al. 2013) and in an evaluation of barcode markers for marine green macroalgae (Saunders and Kucera 2010), this primer had high levels of discriminatory power between species. The second marker, ITS2, has also been widely used in species-level phylogenetic studies of green algae (Lawton et al. 2013).
PCR products were Sanger sequenced by a commercial service (Source Biosciences, Cambridge, UK). Consensus sequences were aligned using the software BioEdit Editor (Hall et al. 2010) and sequences had a higher homology than 98% identity were compared to published data by using NCBI BLAST searches (http://www.ncbi.nlm.nig.gov) (Altschul et al. 1997). Sequence alignments were done by using the Multiple Sequence Comparison by Log-Expectation (MUSCLE) statistical method with the software MEGA X 11.0.11 (http://www.megasoftware.net) (Edgar 2004, Kumar et al. 2018).
RESULTSField collections and algal isolatesWe obtained 345 unialgal macroalgal clones from 308 substrate samples (i.e., an average of 1.12 strains per sample). One hundred and twenty clonal isolates were included in this study. Out of these 120 clones, 84 (70%) were obtained using the GE method, while 35 (29%) were isolated directly from macroscopic thalli collected in the field and 1 strain (0.8%) was obtained in culture by fertilization of fertile cells in vitro (settled zoids) (Supplementary Table S1). Amongst the 120 isolates, 87 (72%) strains were collected in the vicinity of the DE and ZS desalination plants (April 2019) and the other 33 (28%) were collected from different sites on the Kuwait coast (February 2019).
DNA sequences of isolates obtained by Germling EmergenceOf the final 120 isolates selected for molecular identification, the majority were Rhodophyta (n = 51/120 strains; 42%), with the most commonly identified species belonging to the Erythrotrichiaceae G. M. Smith, especially Erythrotrichia Areschoug sp., and Chlorophyta (n = 45/120 strains; 38%) were present in smaller numbers, with many independent replicates of Ulva tepida Y. Masakiyo & S. Shimada (KT374006; described from Australia by Masakiyo and Shimada 2014). Phaeophyceae (n = 24/120 strains; 20%) were present in still smaller numbers, the most frequently identified species being a close relative of Feldmannia mitchelliae (Harvey) H. -S. Kim sensu lato (s.I) (AB302306; described from Japan by Tanaka et al. 2010). Only for 77 out of the 120 isolates, PCR-friendly DNA could be extracted (64%). Of the remaining 43 strains, 17 showed weak PCR amplification which suggested an imperfect match of PCR primers and 14 strains were only identified morphologically (Ulva tepida), but for 12 strains no PCR amplification was achieved.
Based on 130 DNA sequences, using the nuclear ITS2 locus and the chloroplast-encoded tufA primers for Chlorophyta, 26 of the 77 extracted isolates yielding PCR-friendly DNA (corresponding to 34%, representing 17 species) were identified. In the Phaeophyceae, several sequences were obtained of chloroplast psaA, mitochondrial COI, rbcL and RuBisCO spacer (18 of the 77 extracted isolates yielding PCR-friendly DNA, i.e., 23%, representing 14 species). In the Rhodophyta, rbcL and psbA sequences are used to identify 33 out of the 77 isolates with PCR-friendly DNA (43% of the total, representing 26 species). Based on the barcoding results (Supplementary Table S2), 20 sequences supported species-level identity (6 green, 7 brown, and 7 red algae, respectively), 21 enabled genus-level identification, and 14 clustered with higher-level taxonomic entities in each phylum. All sequences were deposited in GenBank/NCBI (outlined in Supplementary Table S2).
DISCUSSIONIn the present work, the GE method with subsequent DNA barcoding to characterize cryptic macroalgal diversity was applied for the first time to seaweeds sampled in the Arabian Gulf region. While this work revealed a number of novel species for the region, the isolated taxa probably underrepresent the diversity of cryptic stages present in the field, as GE is biased towards rapidly growing and proliferating, early successional species.
Diversity and distribution of GE isolates in KuwaitIn the present work, a total of 57 species of algae (Table 2) were identified from 77 DNA-sequenced isolates sampled on the coast of Kuwait, identified morphologically and underpinned by molecular information (Supplementary Table S2). In addition, genetic analyses revealed that the GE isolates sequences were not uniformly distributed among the 3 major macroalgal phyla. Among 120 unialgal isolates, the results revealed 45 isolates representing 17 species of Chlorophyta including Ulvaceae J. V. Lamouroux ex Dumortier, Ulvellaceae Schmidle, Bryopsidaceae Bory, Cladophoraceae Wille, and Planophilaceae Škaloud & Leliaert. Twenty-four isolates representing 14 species in the Sphacelariaceae Decaisne, Acinetosporaceae G. Hamel ex Feldmann, Chordariaceae Greville, Ectocarpaceae C. Agardh, Scytosiphonaceae Farlow, and Bachelotiaceae T. Silberfeld, M.-F. Racault, R. L. Fletcher, A. F. Peters, F. Rousseau & B. de Reviers for Phaeophyceae. According to Peters et al. (2015), species of Chordariaceae are small, with some lacking a macroscopic stage, making it particularly difficult to distinguish species based solely on morphological features. Thus, DNA barcoding has a key role in facilitating species identification in this family. Finally, 51/120 strains coveting Stylonemataceae K. M. Drew, Acrochaetiaceae Melchior, Erythrotrichiaceae G. M. Smith, Spyridiaceae De Toni, Gelidiaceae Kützing, Delesseriaceae Bory, Ceramiaceae Dumortier, Callithamniaceae Kützing, Rhodomelaceae Horaninow, and Bangiaceae Duby represent 26 species of Rhodophyta (Table 2). Overall, 95/120 (79%) of the strains were collected from the Southern Province, including Fintas, whereas 25 (21%) strains were collected from the Northern Province, including Boubiyan Island.
The proportion of Phaeophyceae (n = 18/77 isolates; 23%) was rather small among the initial cultures. Also, species richness among the brown algae isolated here was lower than for the other identified phyla. Amongst the isolates of filamentous Ectocarpales Bessey in the data obtained, Feldmannia Hamel is less well studied than the genus Ectocarpus Lyngbye (Peters et al. 2015), with few reference sequences available in GenBank when using the locus RuBisCO spacer / rbcL region (amplified with primers rbcLP2F, rbcS 952R or rbcL RH3F and rbcS139R). The results obtained in this study showed that the strain K23 [K19-4-130-1] using the 3′-rbcL plus RuBisCO spacer (using primers rbcL1273F, rbcS 139R) was identical (100% homology in both rbcL and RuBisCO spacer) to a sequence for Ectocarpus subulatus from Port Aransas, Texas, USA (accession No. U38750), indicating that this strain belonged to the same species. Originally identified as E. siliculosus, the Ectocarpus from Port Aransas is now regarded as a different species, E. subulatus, which is, in ecophysiological terms, the most low salinity-tolerant species of the genus (Dittami et al. 2020a, 2020b). Port Aransas harbors the southernmost population of Ectocarpus known in North America, with a large temperature amplitude of 17°C between winter (13°) and summer (30°) (Bolton 1983). It is interesting that the same Ectocarpus species appears to be present in Kuwait, where the summer temperature is even higher. Further analyses of the rbcL gene sequences showed that isolates (K72 [K19-4-123-1] and K84 [K19-2-72-5]) had closest matches (98.70 and 98.16% homology) to sequences of Sphacelaria tribuloides Meneghin from the Netherlands (AJ287892) and Okinawa (AJ287891), respectively. The RuBisCO spacer of K84 (262 bp in length) showed 91.6% homology with that of S. tribuloides from Okinawa (AJ287947). Taken together, the results from rbcL and RuBisCO spacer suggest that the material from Kuwait was a closely related species, possibly, to the morphologically similar Sphacelaria novae-hollandiae Sonder, for which no published sequences are available. According to Al-Yamani et al. (2014), both S. tribuloides and S. novae-hollandiae occur in Kuwait. However, for example, Sargassum and Padina sporophytes were not encountered in cultures starting from substrates despite a clear abundance in the field. These algae have no long-lived microstages (microscopic gametophytes or sporophytes) in their life cycle (Peters et al. 2015).
In Kuwait, at the Northern province sites; Boubiyan, Sabiya, Failaka, and Doha, respectively, had less algal diversity than the Southern provience. Three brown algal isolates, Ectocarpus subulatus (K23), Colpomenia sinuosa Derbes et Solier (K87) - Sphacelaria Lyngbye sp. (K71), plus two red taxa, Stylonema alsidii (Zanardini) K. Drew (K12) which has a world-wide geographic distribution (Zuccarello et al. 2011), which had previously been reported from Kuwait (Al-Yamani et al. 2014), and Spyridia Harvey sp. (K66) were identified (Table 3). However, these are clearly adapted to unstable substrates. The limited occurrence of Phaeophyceae and Rhodophyta the Northern Province (Doha) can also be correlated with the muddy substrates there (Jones 1986) which suggests that this area may lack the microstages of taxa in these phyla, compared with other species that were obviously present in this area and that were mostly Chlorophyta. The Northern Province including the Doha site (Table 3) is characterized by shallow, extensive intertidal mud flats with turbid water and semi-enclosed bodies of water with very limited exchange (65 days) to the open sea (Pokavanich and Alosairi 2014) which is typical for Kuwait Bay (Al-Mutairi et al. 2014), compounded by a scarcity of solid, rocky substrates (Alghunaim et al. 2019). The soft nature of these substrates can also explain why the Northern Province is mostly unsuitable for the attachment and growth of several macroalgae particularly large brown macroalgae, such as Sargassum C. Agardh, except for some taxa with small thalli such as Feldmannia sp., which agrees with our observations.
In most temperate coastal waters, macroalgal growth and diversity are directly controlled by nutrient availability (Pedersen et al. 2010, Martínez et al. 2012). The marine environment of Kuwait is also strongly influenced by the discharge of the Shatt Al-Arab estuary with a maximum peak of discharging fresh water from March to July (Al-Said et al. 2017). This freshwater input is associated with a higher concentrations of nutrients, resulting in increased productivity and abundance of green algae in the Doha area. Also, the green algal taxa recorded inhabited calm waters (tidal pools), which were plentiful in the Doha area during winter and early spring where there are many flat, intertidal rock platforms with low current waves, allowing the development of a stable community with higher diversity of green algae (Prathep et al. 2007). Nevertheless, nutrient enrichment and low wave action (Nishihara and Terada 2010) can, therefore, increase the growth of opportunistic seaweeds like green algae characteristically found at Al-Doha, represented in this study by Ulva Linnaeus spp. and members of the Ulvellaceae. It seems that the Ulva species are among the fastest growing macroalgae under conditions of high nutrient and light availability (Phillips and Hurd 2003). Competition among thalli of green algae for space at Doha is intense, in particular when light levels increase in spring and Ulva species generally have high levels of desiccation resistance (Nybakken 1993).
The Chlorophyta recorded in this work (n = 26/77, 34%) mostly have small holdfasts, are smaller and lighter in weight, and are tolerant to the prevailing turbidity and trophic levels compared to Phaeophyceae (Al-Hasan and Jones 1989, Uddin et al. 2011). Furthermore, as Sargassum species are very sensitive to environmental perturbations, substrate type and slope (Fatemi et al. 2012), they are not encountered in the Doha area. A limited number of more tolerant species, however, such as Feldmannia sp. and Iyengaria Børgesen sp., are encountered at Doha. There is some evidence that, over the last two decades, algal diversity on the Doha coast has decreased (Dhia Al-Bader, personal communication / unpublished results).
The Southern Province was the richest area in terms of diversity of algal isolates (95 strains/120; 79%), especially at Fintas and Al-Zour (Table 3). The more benign physical conditions compared to the northern areas of Kuwait probably result in an increase in richness in the “bank” of microscopic stages on this sandy and rocky coast. This finding may be attributed to several factors, such as the lesser anthropogenic impacts on the open sea environment in the south, along with the higher hydrodynamic energy with low turbidity in this region (2 m tidal range) (Al-Yamani et al. 2004). One of the most important factors in species distribution and abundance in tidal areas is exposure to wave action and overall hydrodynamism (Mayakun and Prathep 2005, Nishihara and Terada 2010). According to Al-Yamani et al. (2004), the scattered rocks available on the beaches of this province are suitable for seaweed attachment by holdfasts, resulting in diverse communities of algae. Microscopic stages will inhabit rock surfaces, pebbles and shell fragments, creating a more diverse range of isolates compared to Doha. This finding was emphasized by Al-Yamani et al. (2014) and Al-Hasan and Jones (1989), who also mention this pattern in their previous publications about the macroalgal flora of Kuwait. This is also consistent with Uddin et al. (2011), who mentioned that the southern waters of Kuwait have coral reefs that can also support rich biodiversity of marine macrophytes. Al-Zour is characterized by a rocky shore and overall is a protected and sheltered area, therefore a high diversity of seaweeds, especially among the Rhodophyta, was not surprising. Overall, in the present study, the Rhodophyta were the phylum with the highest species richness compared to the green and brown phyla among seaweeds isolated and identified in this study, constituting 42% (n = 51/120) of the total algal isolates, representing 26 species. These observations are consistent with several previous reports (Silva et al. 1996, Price et al. 2006, John 2012, John and Al-Thani 2014) who suggested that among a total of approximately 282 benthic marine algal species in the Arabian Gulf, the most diverse were the Rhodophyta (147 spp.).
A further exploration of the macroalgal diversity on the coast of Kuwait will likely reveal greater diversity, especially of smaller epiphytic algal species with short life cycles, as the time period covered in this work encompassed only late winter (February 2019) and spring (April 2019). The GE method was clearly a good tool to investigate cryptic macroalgal floral diversity. Compared to environmental sequencing (metabarcoding) it offers the advantage of providing access to a plethora of biological information such as morphology, life cycle, physiology and biochemistry. This method has substantially helped revealing a diversity in the local Kuwaiti waters that was not previously known. This study has revealed several new records of species and genera, including noteworthy are new records of Porphyrostromium, a new genus from the Bangiales namely Kuwaitiella rubra gen. et sp. nov., and two unclear species for Planophilaceae. The data obtained in this present study will also help to predict the future of algal diversity and floristic composition under ongoing and predicted climate change effects impacting this already harsh environment.
ACKNOWLEDGEMENTSThe present work formed part of the first author’s PhD thesis ‘Macroalgal biodiversity of Kuwait, with special emphasis on the vicinity of desalination plants’. We acknowledge Dr. Hedda Weitz (University of Aberdeen) for providing help in the laboratory and from Ioanna Kosma (University of the Aegean) and Andreas Henkel (Kuwait University) for diving and logistics support during the expedition to Kuwait. We acknowledge the funding received to support this work from the Marine Alliance for Science and Technology (grant reference HR09011) to FCK and Kuwait Foundation for the Advancement of Science (KFAS; grant number PR17125L18) to DA. To Mr. Yusuf Buhadi from the department of Marine Sciences at Kuwait University for his help in the field work and to Mrs. Nisha V. S. Vadakkhancheril for photography.
SUPPLEMENTARY MATERIALSSupplementary Table S1Identification, habitat, and collection sites of the 120 algal cultures in Kuwait (https://www.e-algae.org). Supplementary Table S2List of isolated algal strains collected from Kuwait, grouped according to the closest sequence match in GenBank (https://www.e-algae.org) Supplementary Fig. S1(A–C) The identification of the species was based upon molecular analyses of in vitro algal cultures (under laboratory conditions with specific light : dark time period)—mostly in a microscopic stage and not always similar morphologically to growth in the natural environment (https://www.e-algae.org). Fig. 1Localities and sites sampled. (A) General location of Kuwait in the northwestern of the Arabian Gulf. (B) Map of Kuwait represents 1–12 sampling points including shoreline and off shore Islands namely; Bubiyan, Sabiya, Failaka, Doha, Kuwait City Sharq, Fintas, Kubbar, Qaruh, Al- Zour, Al-Khiran, Umm Al-Maradim, and Nuwaiseeb, respectively. Subset of maps showing sample locations at (C) Doha 4 and (D) Al-Zour 9 and Al- Khiran 10 sites (GIS maps produced using ArcGIS software; National Geographic, Esri, Garmin, USA). 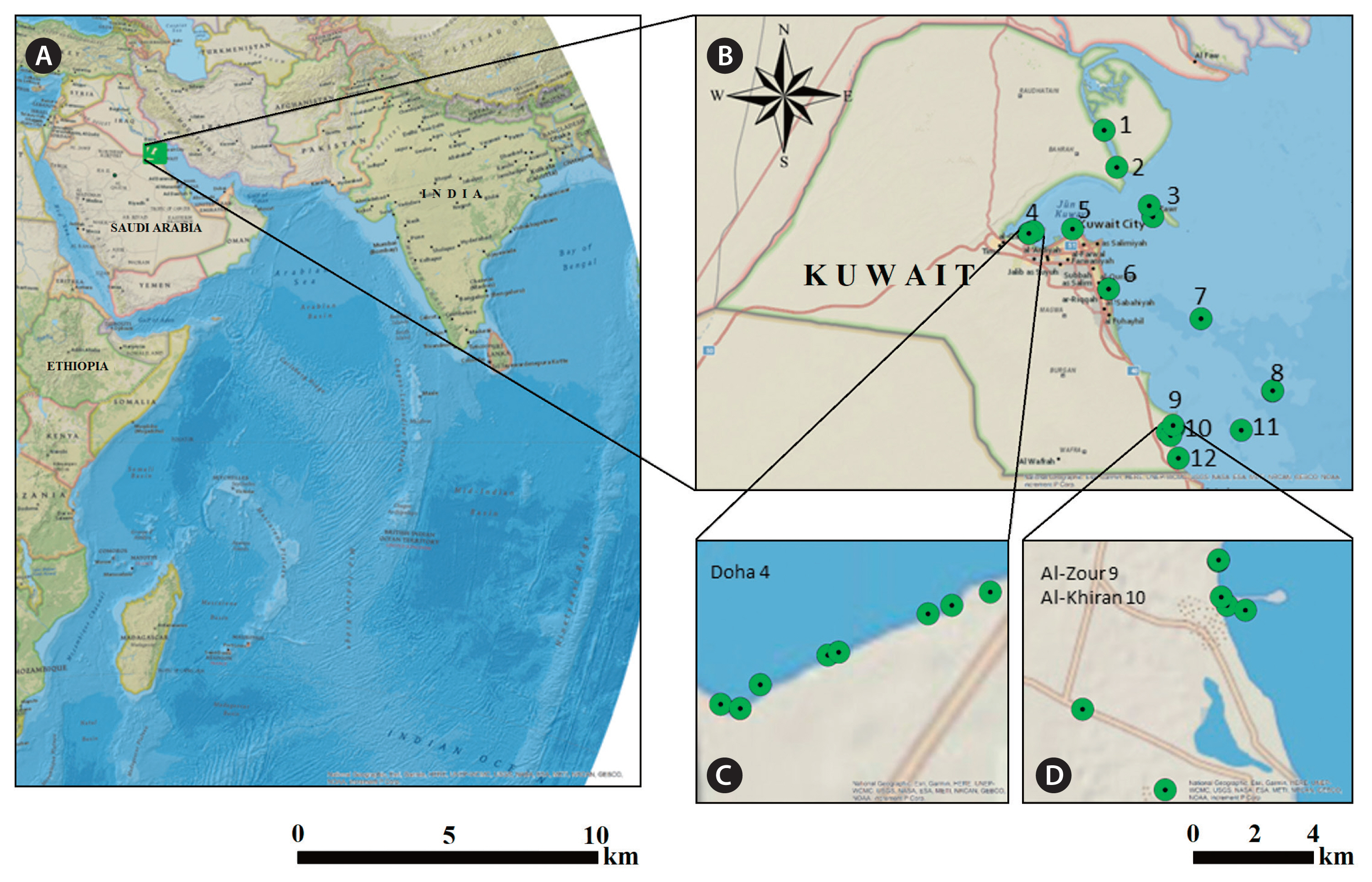
Table 1List of oligonucleotide primers of amplification used to amplify different fragments of DNA from genomes of algal isolates
Table 2Species names and authorities for Chlorophyta, Phaeophyceae, and Rhodophyta isolated from Kuwait (February–April 2019) based on DNA barcoding results
Table 3Diversity of algal isolates obtained by Germling Emergence Method based on 90 algal cultures collected from Kuwaiti costal sites REFERENCESAl-Adilah, H., Al-Bader, D., Elktob, M., Kosma, I., Kumari, P. & Küpper, F. C. 2021. Trace element concentrations in seaweeds of the Arabian Gulf identified by morphology and DNA barcodes. Bot. Mar. 64:327–338.
Al-Adilah, H., Peters, A. F., Al-Bader, D., Raab, A., Akhdhar, A., Feldmann, J. & Küpper, F. C. 2020. Iodine and fluorine concentrations in seaweeds of the Arabian Gulf identified by morphology and DNA barcodes. Bot. Mar. 63:509–519.
Alghunaim, A., Taqi, A., Al-Kandari, M. & Al-Said, T. 2019. Distribution and nature of Sargassum species in the Kuwait waters. J. Geosci. Environ. Prot. 7:53–59.
Al-Hasan, R. H. & Jones, W. E. 1989. Marine algal flora and sea grasses of the coast of Kuwait. J. Univ. Kuwait Sci. 16:289–341.
Al-Mutairi, N., Abahussain, A. & Al-Battay, A. 2014. Environmental assessment of water quality in Kuwait Bay. Int. J. Environ. Sci. Dev. 5:527–532.
Al-Said, T., Al-Ghunaim, A., Subba Rao, D. V., Al-Yamani, F., Al-Rifaie, K. & Al-Baz, A. 2017. Salinity-driven decadal changes in phytoplankton community in the NW Arabian Gulf of Kuwait. Environ. Monit. Assess. 189:268 pp.
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402.
Al-Yamani, F. Y., Bishop, J., Ramadhan, E., Al-Husain, M. & Al-Ghadban, A. N. 2004. Oceanographic atlas of Kuwait waters. Kuwait Institute for Scientfic Research, Kuwait, 203 pp.
Al-Yamani, F. Y., Polikarpov, I., Al-Ghunaim, A. & Mikhaylova, T. 2014. Field guide of marine macroalgae (Chlorophyta, Rhodophyta, Phaeophyceae) of Kuwait. Kuwait Institute for Scientific Research, Kuwait, 190 pp.
Bartolo, A. G., Zammit, G., Peters, A. F. & Küpper, F. C. 2020. The current state of DNA barcoding of marcroalgae in the Mediterranean Sea: presently lacking but urgently required. Bot. Mar. 63:253–272.
Bolton, J. J. 1983. Ecoclinal variation in Ectocarpus siliculosus (Phaeophyceae) with respect to temperature growth optima and survival limits. Mar. Biol. 73:131–138.
Broom, J. E. S., Nelson, W. A., Farr, T. J., Phillips, L. E. & Clayton, M. 2010. Relationships of the Porphyra (Bangiales, Rhodophyta) flora of the Falkland Islands: a molecular survey using rbc L and nSSU sequence data. Aust. Syst. Bot. 23:27–37.
Coelho, S. M., Scornet, D., Rousvoal, S., Peters, N. T., Dartevelle, L., Peters, A. F. & Cock, J. M. 2012.
Ectocarpus: a model organism for the brown algae. Cold Spring Harb. Protoc. 2012. 193–198.
Destombe, C. & Douglas, S. E. 1991. Rubisco spacer sequence divergence in the rhodophyte alga Gracilaria verrucosa and closely related species. Curr. Genet. 19:395–398.
De Vargas, C., Audic, S., Henry, N., Decelle, J., Mahé, F., Logares, R., Lara, E., Berney, C., Le Bescot, N., Probert, I., Carmichael, M., Poulain, J., Romac, S., Colin, S., Aury, J.-M., Bittner, L., Chaffron, S., Dunthorn, M., Engelen, S., Flegontova, O., Guidi, L., Horák, A., Jaillon, O., Lima-Mendez, G., Lukeš, J., Malviya, S., Morard, R., Mulot, M., Scalco, E., Siano, R., Vincent, F., Zingone, A., Dimier, C., Picheral, M., Searson, S., Kandels-Lewis, S., Acinas, S. G., Bork, P., Bowler, C., Gorsky, G., Grimsley, N., Hingamp, P., Iudicone, D., Not, F., Ogata, H., Pesant, S., Raes, J., Sieracki, M. E., Speich, S., Stemmann, L., Sunagawa, S., Weissenbach, J., Wincker, P.; Karsenti, E. & Tara Oceans Coordinators 2015. Eukaryotic plankton diversity in the sunlit ocean. Science. 348:1261605 pp.
Dittami, S. M., Corre, E., Brillet-Guéguen, L., Lipinska, A.P., Pontoizeau, N., Aite, M., Avia, K., Caron, C., Cho, C.H., Collén, J., Cormier, A., Delage, L., Doubleau, S., Frioux, C., Gobet, A., González-Navarrete, I., Groisillier, A., Herv, C., Jollivet, D., KleinJan, H., Leblanc, C., Liu, X., Marie, D., Markov, G. V., Minoche, A. E., Monsoor, M., Pericard, P., Perrineau, M. M., Peters, A. F., Siegel, A., Siméon, A., Trottier, C., Yoon, H. S., Himmelbauer, H., Boyen, C. & Tonon, T. 2020a. The genome of Ectocarpus subulatus: a highly stress-tolerant brown alga. Mar. Genomics. 52:100740 pp.
Dittami, S. M., Peters, A. F., West, J. A., Cariou, T., KleinJan, H., Burgunter-Delamare, B., Prechoux, A., Egan, E. & Boyen, C. 2020b. Revisiting Australian Ectocarpus subulatus (Phaeophyceae) from the Hopkins River: distribution, abiotic environment, and associated microbiota. J. Phycol. 56:719–729.
Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797.
Famà, P., Wysor, B., Kooistra, W. H. C. F. & Zuccarello, G. C. 2002. Molecular phylogeny of the genus Caulerpa (Caulerpales, Chlorophyta) inferred from chloroplast tuf A gene. J. Phycol. 38:1040–1050.
Fatemi, S. M. R., Ghavam Mostafavi, P., Rafiee, F. & Saeed Taheri, M. 2012. The study of seaweeds biomass from intertidal rocky shores of Qeshm Island, Persian Gulf. Int. J. Mar. Sci. Environ. 2:101–106.
Gachon, C. M. M., Strittmatter, M., Müller, D. G., Kleinteich, J. & Küpper, F. C. 2009. Detection of differential host susceptibility to the marine oomycete pathogen Eurychasma dicksonii by real-time PCR: not all algae are equal. Appl. Environ. Microbiol. 75:322–328.
Hall, J. D., Fučíkov, K., Lo, C., Lewis, L. A. & Karol, K. G. 2010. An assessment of proposed DNA barcodes in freshwater green algae. Cryptogam. Algol. 31:529–555.
Han, K. Y., Graf, L., Reyes, C. P., Melkonian, B., Andersen, R. A., Yoon, H. S. & Melkonian, M. 2018. A re-investigation of Sarcinochrysis marina (Sarcinochrysidales, Pelagophyceae) from its type locality and the descriptions of Arachnochrysis, Pelagospilus, Sargassococcus and Sungminbooa genera nov. Protist. 169:79–106.
Hasan, A. H., Van der Aa, P., Küpper, F. C., Al-Bader, D. & Peters, A. F. 2022.
Kuwaitiella rubra gen. et sp. nov. (Bangiales, Rhodophyta), a new filamentous genus and species from the north-western Indian Ocean. Phycol. Res. 70:192–202.
Hodge, F. J., Buchanan, J. & Zuccarello, G. C. 2010. Hybridization between the endemic brown algae Carpophyllum maschalocarpum and Carpophyllum angustifolium (Fucales): genetic and morphological evidence. Phycol. Res. 58:239–247.
Hoffmann, A. J. & Santelices, B. 1991. Banks of algal microscopic forms: hypotheses on their functioning and comparisons with seed banks. Mar. Ecol. Prog. Ser. 79:185–194.
John, D. M. 2012. Marine algae (seaweeds) associated with coral reefs of the Gulf. In : Riegl B. M., Purkis S. J., editors Coral Reefs of the Gulfs: Adaptation to Climate Extremes. Coral Reefs of the World. 3:Springer, Dordrecht, 170–186.
John, D. M. & Al-Thani, R. F. 2014. Benthic marine algae of the Arabian Gulf: a critical review and analysis of distribution and diversity patterns. Nova Hedwigia. 98:341–392.
Jones, D. 1986. A field guide to the seashores of Kuwait and the Arabian Gulf. University of Kuwait, Kuwait, 193 pp.
Kawai, H., Hanyuda, T., Draisma, S. G. A. & Müller, D. G. 2007. Molecular phylogeny of Discosporangium mesarthrocarpum (Phaeophyceae) with a reinstatement of the order discosporangiales. J. Phycol. 43:186–194.
Kirkendale, L., Saunders, G. W. & Winberg, P. 2013. A molecular survey of Ulva (Chlorophyta) in temperate Australia reveals enhanced levels of cosmopolitanism. J. Phycol. 49:69–81.
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35:1547–1549.
Kunimoto, M., Kito, H., Kaminishi, Y., Mizukami, Y. & Murase, N. 1999. Molecular divergence of the SSU rRNA gene and internal transcribed spacer 1 in Porphyra yezoensis (Rhodophyta). J. Appl. Phycol. 11:211–216.
Küpper, F. C., Peters, A. F., Shewring, D. M., Sayer, M. D. J., Mystikou, A., Brown, H., Azzopardi, E., Dargent, O., Strittmatter, M., Brennan, D., Asensi, A. O., van West, P. & Wilce, R. T. 2016. Arctic marine phytobenthos of northern Baffin Island. J. Phycol. 52:532–549.
Lane, C. E., Mayes, C., Druehl, L. D. & Saunders, G. W. 2006. A multi-gene molecular investigation of the kelp (Laminariales, Phaeophyceae) supports substantial taxonomic re-organization. J. Phycol. 42:493–512.
Lawton, R. J., Mata, L., de Nys, R. & Paul, N. A. 2013. Algal bioremediation of waste waters from land-based aquaculture using Ulva: selecting target species and strains. PLoS ONE. 8:e77344 pp.
Lewin, J. 1966. Silicon metabolism in diatoms. V. Germanium dioide, a specific inhibitor of diatom growth. Phycologia. 6:1–12.
Macreadie, P. I., Jarvis, J., Trevathan-Tackett, S. M. & Bellgrove, A. 2017. Seagrasses and macroalgae: importance, vulnerability and impacts. In : Phillips B. F., Pérez-Ramírez M., editors Climate Change Impacts on Fisheries and Aquaculture: A Global Analysis. Wiley-Blackwell, Oxford, 729–770.
Martínez, B., Pato, L. S. & Rico, J. M. 2012. Nutrient uptake and growth responses of three intertidal macroalgae with perennial, opportunistic and summer-annual strategies. Aquat. Bot. 96:14–22.
Masakiyo, Y. & Shimada, S. 2014. Species diversity of the genus Ulva (Ulvophyceae, Chlorophyta) in Japanese waters, with special reference to Ulva tepida Masakiyo et S. Shimada sp. nov. Bull. Natl. Mus. Nat. Sci. Ser. B Bot. 40:1–13.
Mayakun, J. & Prathep, A. 2005. Seasonal variations in diversity and abundance of macroalgae at Samui Island, Surat Thani Province, Thailand. Songklanakarin J. Sci. Technol. 27:653–663.
Müller, D. G. & Ramírez, M. E. 1994. Filamentous brown algae from the Juan Fernandez Archipelago (Chile): contribution of laboratory culture techniques to a phytogeographic survey. Bot. Mar. 37:205–211.
Nishihara, G. N. & Terada, R. 2010. Spatial variations in nutrient supply to the red algae Eucheuma serra (J. Agardh) J. Agardh. Phycol. Res. 58:29–34.
Nybakken, J. W. 1993. Marine biology an ecological approach. Harper Collins College Publishers, NY, 10022 pp.
Pedersen, M. F., Borum, J. & Fotel, F. L. 2010. Phosphorus dynamics and limitation of fast- and slow-growing temperate seaweeds in Oslofjord, Norway. Mar. Ecol. Prog. Ser. 399:103–115.
Peters, A. F., Couceiro, L., Tsiamis, K., Küpper, F. C. & Valero, M. 2015. Barcoding of cryptic stages of marine brown algae isolated from incubated substratum reveals high diversity in Acinetosporaceae (Ectocarpales, Phaeophyceae). Cryptogam. Algol. 36:3–29.
Peters, A. F. & Ramírez, M. E. 2001. Molecular phylogeny of small brown algae, with special reference to the systematic position of Caepidium antarcticum (Adenocystaceae, Ectocarpales). Cryptogam. Algol. 22:187–200.
Peters, A. F., Scornet, D., Müller, D. G., Kloareg, B. & Cock, J. M. 2004. Inheritance of organelles in artificial hybrids of the isogamous multicellular chromist alga Ectocarpus siliculosus (Phaeophyceae). Eur. J. Phycol. 39:235–242.
Phillips, J. C. & Hurd, C. L. 2003. Nitrogen ecophysiology of intertidal seaweeds from New Zealand: N uptake, storage and utilisation in relation to shore position and season. Mar. Ecol. Prog. Ser. 264:31–48.
Pirian, K., Piri, K., Sohrabipour, J., Jahromi, S. T. & Blomster, J. 2016. Molecular and morphological characterisation of Ulva chaugulii, U. paschima and U. ohnoi (Ulvophyceae) from the Persian Gulf, Iran. Bot. Mar. 59:147–158.
Pokavanich, T. & Alosairi, Y. 2014. Summer flushing characteristics of Kuwait Bay. J. Coast. Res. 30:1066–1073.
Prathep, A., Wichachucherd, B. & Thongroy, P. 2007. Spatial and temporal variation in density and thallus morphology of Turbinaria ornata in Thailand. Aquat. Bot. 86:132–138.
Price, A. R. G., Vincent, L. P. A., Venkatachalam, A. J., Bolton, J. J. & Basson, P. W. 2006. Concordance between different measures of biodiversity in Indian Ocean macroalgae. Mar. Ecol. Prog. Ser. 319:85–91.
Provasoli, L. 1966. Media and prospects for the cultivation of marine algae. In : Watanabe A., Hattori A., editors Cult. Collect. Algae: Proc. US-Japan Conf. Japan Society of Plant Physiology, Hakone, 63–75.
Ramírez, M. E. & Müller, D. G. 1991. New records of benthic marine algae from Easter Island. Bot. Mar. 34:133–137.
Rizouli, A., Küpper, F. C., Louizidou, P., Mogg, A. O. M., Azzopardi, E., Sayer, M. D. J., Kawai, H., Hanyuda, T. & Peters, A. F. 2020. The minute alga Schizocladia ischiensis (Schizocladiophyceaase, Ochrophyta) isolated by germling emergence from 24 m depth of Rhodes (Greece). Diversity. 12:102 pp.
Santiañez, W. J. E., Al-Bader, D., West, J. A., Bolton, J. J. & Kogame, K. 2020. Status, morphology, and phylogenetic relationships of Iyengaria (Scytosiphonaceae, Phaeophyceae), a brown algal genus with a disjunct distribution in the Indian Ocean. Phycol. Res. 68:323–331.
Saunders, G. W. 2005. Applying DNA barcoding to red macroalgae: a preliminary appraisal holds promise for future applications. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360:1879–1888.
Saunders, G. W. & Kucera, H. 2010. An evaluation of rbcL, tufA, UPA, LSU and ITS as DNA barcode markers for the marine green macroalgae. Cryptogam. Algol. 31:487–528.
Saunders, G. W. & McDevit, D. C. 2013. DNA barcoding unmasks overlooked diversity improving knowledge on the composition and origins of the Churchill algal flora. BMC Ecol. 13:9 pp.
Schoenrock, K. M., McHugh, T. A. & Krueger-Hadfield, S. A. 2021. Revisiting the ‘bank of microscopic forms’ in macroalgal-dominated ecosystems. J. Phycol. 57:14–29.
Siemer, B. L., Stam, W. T., Olsen, T. J. & Pedersen, P. M. 1998. Phylogenetic relationships of the brown algal orders Ectocarpales, Chordariales, Dictyosiphonales, and Tilopteridales (Phaeophyceae) based on RUBISCO large subunit and spacer sequences. J. Phycol. 34:1038–1048.
Silva, P. C., Basson, P. W. & Moe, R. L. 1996. Catalogue of the benthic marine algae of the Indian Ocean. 79:University California Press, Berkeley, CA, 1259 pp.
Starr, R. C. & Zeikus, J. A. 1993. UTEX: the culture collection of algae at the University of Texas at Austin 1993 List of cultures. J. Phycol. 29:1–106.
Tanaka, A., Uwai, S., Nelson, W. & Kawai, H. 2010.
Phaeophysema gen. nov. and Vimineoleathesia gen. nov., new brown algal genera for the minute Japanese members of the genus Leathesia
. Eur. J. Phycol. 45:107–115.
Tarakhovskaya, E. R., Kang, E. J., Kim, K. Y. & Garbary, D. J. 2012. Effect of GeO2 on embryo development and photosynthesis in Fucus vesiculosus (Phaeophyceae). Algae. 27:125–134.
Uddin, S., Al Ghadban, A. N. & Khabbaz, A. 2011. Localized hyper saline waters in Arabian Gulf from desalination activity: an example from South Kuwait. Environ. Monit. Asses. 181:587–594.
West, J. A., Loiseaux-De Goër, S. & Zuccarello, G. C. 2012. Upright Erythropeltidales (Rhodophyta) in Brittany, France and description of a new species, Erythrotrichia longistipitata
. Cah. Biol. Mar. 53:255–270.
White, T. J., Bruns, T., Lee, S. & Taylor, J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In : Innis M. A., Gelfand D. H., Sninsky J. J., White T. J., editors PCR Protocols: A Guide to Methods and Applications. Academic Press, San Diego, CA, 315–322.
Wolf, M. 2012. Molecular and morphological investigations on seaweed biodiversity and alien introductions in the Adriatic Sea (Mediterranean, Italy). Ph.D. dissertation. Universit Degli Studi Di padova Dipartimento Di Biologia, Padova, Italy, 183 pp.
Wynne, M. J., Kamiya, M., West, J.A., Loiseaux-de Goër, S., Lim, P.-E., Sade, A., Russell, H. & Küpper, F. C. 2020. Morphological and molecular evidence for the recognition of Hypoglossum sabahense sp. nov. (Delesseriaceae, Rhodophyta) from Sabah, Malaysia. Algae. 35:157–165.
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||