INTRODUCTION
Cryptic species are defined as two or more distinct entities classified as a single nominal species owing to their superficial morphological similarity (Bickford et al. 2007). These taxa result from the occurrence of distinct evolutionary lineages within species (Struck et al. 2018). In addition, genetic variation can be used to presume species adaptation during environmental changes because species with high genetic variation often have a reduced risk of extinction (Benson et al. 2022). Despite the advances in DNA sequencing and molecular techniques to detect them, algal taxa with potential cryptic diversity remain uncovered. Therefore, taxonomists have made steady efforts to reveal cryptic species and investigated the genetic variation in populations of those species. Several studies have reported various cryptic species among green (Lee et al. 2019), brown (Vieira et al. 2022), and red seaweeds (Muangmai et al. 2022). One interesting example of cryptic diversity is Gloiopeltis furcata (Postels & Ruprecht) J. Agardh, a red algal species in which 11 distinct cryptic lineages were identified (Yang et al. 2020, 2021a). Such unexpected cryptic diversity within a species detected using molecular analysis stimulated interest in other red algal species that are difficult to identify based on morphological characteristics alone.
Plocamium Lamouroux is a red algal genus distributed worldwide, from Antarctica to the Arctic, with 46 accepted species names (Guiry and Guiry 2023). Species of Plocamium are mainly characterized by the number of branchlets (ramuli) in alternating series, lower branchlet morphology, tetrasporangial stichidia structures, and the arrangement of cystocarps (Womersley 1994, Wynne 2002). However, species identification is challenging owing to environmentally driven morphological plasticity (Yano et al. 2004, Saunders and Lehmkuhl 2005). Some species of Plocamium contain halogenated monoterpenes with anticancer and insecticidal properties (Rovirosa et al. 2013, Sabry et al. 2017), while other species from Antarctica have the most common halogenated monoterpenes with the highest secondary metabolite diversity among and within populations as well as among individual (Shilling et al. 2019, 2021). Portieria Zanardini, another red algal genus, has differences in secondary metabolites between separate life history stages, suggesting a correlation between haplotypic diversity (Payo et al. 2011, Yang and Kim 2018). Thus, molecular studies of genetic variation are essential for clear species recognition, particularly for species containing these functional biomolecules, and for discovering hotspots for species conservation.
Plocamium cartilagineum (Linnause) P.S. Dixon, which type locality is known as northern Europe, has been reported worldwide (Guiry and Guiry 2023), including in the Northwest (NW) Pacific region, encompassing Korea, Japan, and China. This species is characterized by having alternating ramuli in a series of 2–6, simple tetrasporangial stichidia, and sessile cystocarps. However, previous studies revealed considerable morphological plasticity in P. cartilagineum (Saunders and Kraft 1994). Furthermore, molecular phylogenetic analyses indicated that specimens referred to as P. cartilagineum based on morphology may represent cryptic diversity from different geographic locations (Saunders and Lehmkuhl 2005, Cremades et al. 2011). Saunders and Lehmkuhl (2005) discovered eight genetic cryptic species in P. cartilagineum specimens worldwide using the partial large subunit (LSU) ribosomal RNA (rRNA) gene, which provided unequivocal evidence that this species is not cosmopolitan in distribution. To resolve the taxonomic ambiguity of P. cartilagineum among European specimens, Cremades et al. (2011) revised the taxonomic and nomenclatural proposal for the molecular entity of this species using 5′ region of cytochrome c oxidase subunit I (COI-5P), suggesting a new interpretation for P. cartilagineum sensu stricto (EUR1) and P. subtile Kützing (EUR2), previously recognized by Saunders and Lehmkuhl (2005). Both studies suggested that specimens in the NW Pacific region, including Korea and Japan, may represent a new cryptic species genetically differentiated but morphologically indistinguishable from European specimens (Saunders and Lehmkuhl 2005, Cremades et al. 2011).
Plocamium telfairiae (W. J. Hooker & Harvey) Harvey ex Kutzing has been reported based on morphological identification from the western Indian Ocean to the NW Pacific Ocean, and its type locality is Mauritius (Guiry and Guiry 2023). De Clerck et al. (2002) reported this species growing in intertidal rock pools and sublittoral fringes at a depth of 15 m in northern Kwazulu-Natal, South Africa. They described its alternately-distichously branched ramuli and tetrasporangial stichidia formed on the axils of the ramuli. However, phylogenetic studies suggest that this South African species differs from specimens from Korea and Japan (Saunders and Lehmkuhl 2005, Cremades et al. 2011, Reddy et al. 2023). Saunders and Lehmkuhl (2005) stated that Plocamium specimens in the NW Pacific region require further investigation, indicating that P. telfairiae may also contain cryptic diversity.
In Korea, four Plocamium taxa, P. cartilagineum, P. ovicorne Okamura, P. telfairiae, and P. telfairiae f. uncinatum Okamura, have been reported (Kim and Hwang 2015). They are found in intertidal and subtidal waters on the eastern to southern coasts, including Jeju Island (Kang and Kim 2012). The antioxidant activity of polyphenols in P. telfairiae from Korea has been studied. The extract of this species is also used in functionalized Au nanostructures and reported to possess anti-adipogenic activity (Kim et al. 2007, Park et al. 2022). For species conservation and utilization of the bioactive substances in this genus, precise species identification and research on hotspots with high genetic diversity, respectively, are required. However, no in-depth phylogeographic studies of Plocamium species have been conducted in Korea, unlike the studies on medical uses of extracts in this genus.
Hotspots, which are areas with high intraspecific genetic diversity, are important for species conservation (Chiocchio et al. 2021), as they contain populations that can adapt to selective pressures (Zampiglia et al. 2019, Benson et al. 2022). However, the spatial patterns of genetic variation in the genus Plocamium are insufficient to detect priority areas for conservation in the NW Pacific. To uncover cryptic diversity and hotspots with high genetic variation in this genus in the NW Pacific region, it is necessary to obtain information on accurate species delimitation and the genetic variation of each species based on molecular analyses. Therefore, this study aims (1) to identify potential cryptic diversity analyzing the phylogenetic affinities for specimens, known as P. cartilagineum and P. telfairiae from the NW Pacific and their type localities, (2) to characterize the level of genetic variation in two Plocamium species, using COI-5P and rbcL genes, and (3) to find hotspots with high intraspecific genetic diversity through the phylogeographic study of Plocamium species in the NW Pacific region.
MATERIALS AND METHODS
Samples and observations
A total of 209 samples of Plocamium spp. were obtained from 14 localities in Korea and three in Japan (Supplementary Table S1). Intertidal sampling was conducted during low tide, whereas subtidal sampling was conducted by SCUBA diving. In the field, the samples were first identified as P. “telfairiae” and P. “cartilagineum” based on morphological traits (Kim and Hwang 2015). A portion of each branch was excised from plants and desiccated using silica gel for DNA analyses. For morphological observations, specimens were preserved in 4% formalin in seawater. Sections were manually prepared using a razor or a bench-top freezing microtome (NK-101-II; Nippon Optical Works Co, Ltd., Tokyo, Japan). Photomicrographs were obtained using a BX43 microscope (Olympus, Tokyo, Japan) equipped with an EOS 600D digital camera (Canon, Tokyo, Japan). Digitized images were adjusted for clarity using Adobe Photoshop software (ver. 6.1; Adobe Systems Inc., San Jose, CA, USA). Voucher specimens were deposited in the herbarium of the Jeju National University (JNUB), Jeju, Korea.
DNA extraction, amplification, and sequencing
Genomic DNA was extracted using a MagPurix DNA Isolation Kit (Zinext, Taipei, Taiwan), according to the manufacturer’s instructions. The extracted DNA was stored at −20°C and subjected to polymerase chain reaction (PCR) amplification using AccuPower PCR Premix (Bioneer, Daejeon, Korea) at a final volume of 20 μL. Targeted sequences of the COI-5P and rbcL genes were amplified and sequenced using GazF2 and GazR1 primers for COI-5P (Saunders 2005, Lane et al. 2007) and F7–R898 and F762–R1442 for rbcL (Kim et al. 2010). PCR conditions were as described by Yang et al. (2021b). PCR products were visually examined on a 1% agarose gel, purified using ExoSAP-IT (USB, Cleveland, OH, USA), and commercially sequenced (Macrogen Inc., Seoul, Korea).
Alignment, genetic diversity, and haplotype analyses
All successful amplifications for COI-5P and rbcL genes were sequenced in both directions. Assembly and manual editing were performed using Geneious Prime ver. 2022.2.2 (http://www.geneious.com). All edited sequences were combined with available GenBank sequences and aligned using the MUSCLE algorithm in Geneious. DNA polymorphism levels of COI-5P were summarized for two Plocamium species in the NW Pacific, in terms of the number of haplotype (Nh), haplotype diversity (H), and nucleotide diversity (π) using Arlequin ver. 3.5.2 (Excoffier et al. 2005). To evaluate the relationships among COI-5P haplotypes, a haplotype network was constructed with a minimum spanning network in Arlequin. Differences in the genetic structure of populations based on COI-5P were assessed computing pairwise FST statistics in Arlequin between regions. Uncorrected p-distances implemented in MEGA X (Kumar et al. 2018) were used to calculate genetic divergence in terms of pairwise differences between and within species.
Species delimitation and phylogenetic analyses
For the phylogenetic tree reconstruction, Sarcodia diliata was used as the outgroup, and a sequence of each haplotype of Plocamium spp. was used for both genes. Maximum likelihood analysis was performed using the General Time Reversible GAMMA substitution model with 1,000 bootstrap supports in Geneious. Bayesian inference was performed using MrBayes v.3.2.1 (Ronquist et al. 2012) using the Metropolis-coupled Markov Chain Monte Carlo (MCMC) model. Four million generations of two independent runs were performed with four chains, sampling trees for every 100 generations. The burn-in period was graphically identified by tracking the likelihood of each generation to determine whether they reached a plateau. The first 25% of saved trees were removed, and the remaining trees were used to calculate Bayesian posterior probabilities.
To evaluate species delineation, three molecular delimitation analyses were applied to the COI-5P and rbcL datasets: Assemble Species by Automatic Partitioning (ASAP) (Puillandre et al. 2021); generalized mixed Yule-coalescent (GMYC) (Fujisawa and Barraclough 2013); and Poisson Tree Process (bPTP) (Zhang et al. 2013). ASAP analysis was performed through a web-based interface (https://bioinfo.mnhn.fr/abi/public/asap), using default settings for JC69 and K2P. ASAP is a new method for building species partitions from single-locus sequence alignments based on pairwise genetic distance and provides a score for each defined partition (Puillandre et al. 2021). The GMYC method requires an ultrametric tree, which was obtained using Bayesian analyses in BEAST v.1.10.4 (Suchard et al. 2018), with divergence times estimated under an uncorrelated lognormal relaxed molecular clock model (Drummond et al. 2012) and the Yule-process as the prior tree. The Bayesian MCMC was run for 20 million generations, with trees and parameters sampled every 100 generations. The output was assessed for convergence using Tracer v.1.7.1 (Rambaut et al. 2018). After removing 25% of the trees as burn-in, the remaining trees were used to generate a single summarized tree in TreeAnnotator v.1.10.4 (part of the BEAST v.1.10.4 package) as an input file for the GMYC analyses. GMYC analyses with a single threshold model were performed using R software (R Development Core Team, https://www.R-project.org) under the “splits” package using the “gmyc” function (R-Forge, https://r-forge.r-project.org/projects/splits/). bPTP analysis was performed using a bifurcated phylogenetic input tree, as implemented in the bPTP web server (https://species.h-its.org). The parameters were set as follows: 500,000 MCMC generations, thinning by a factor of 100, and a 10% burn-in.
RESULTS
Phylogenetic analyses
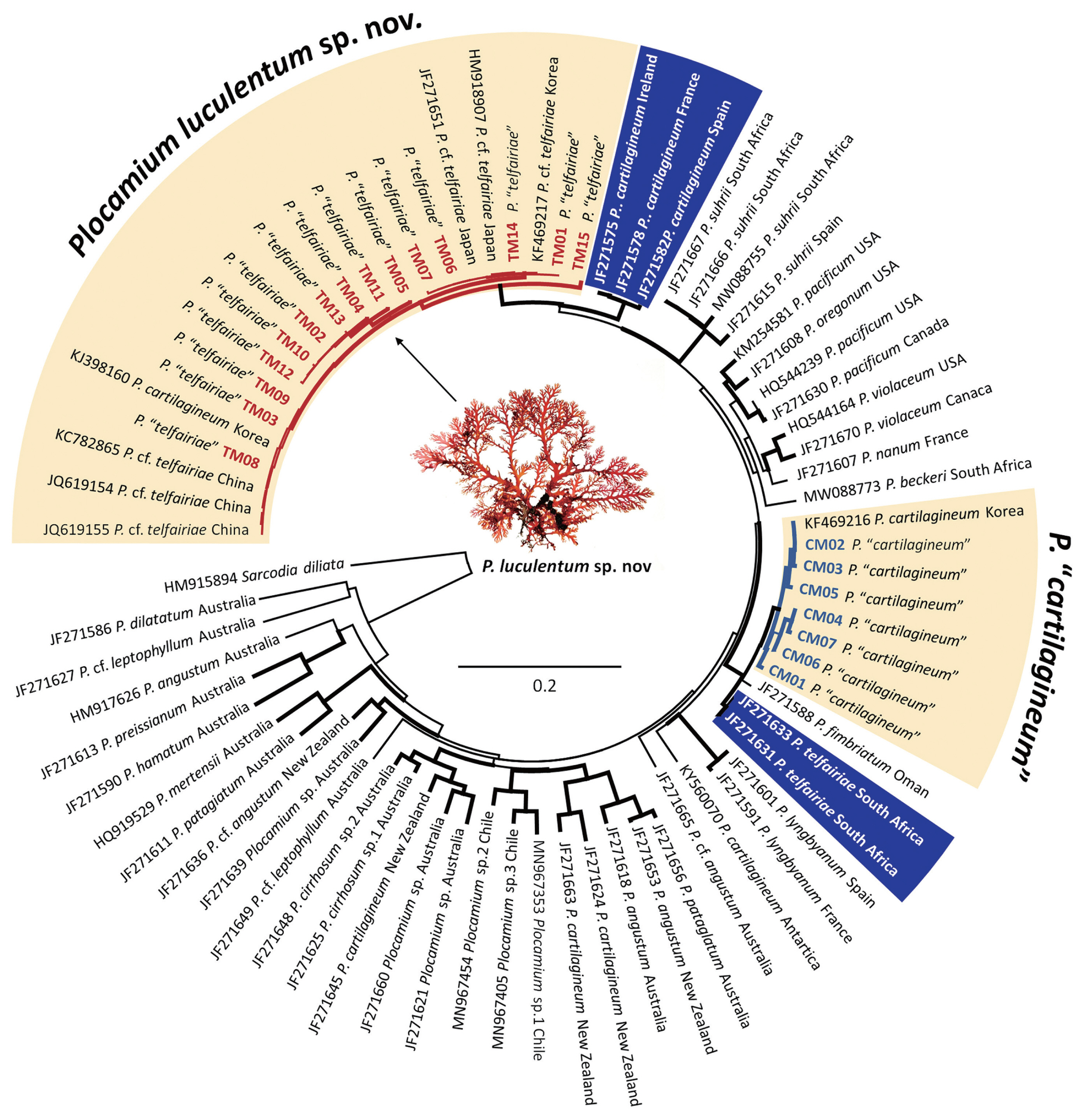
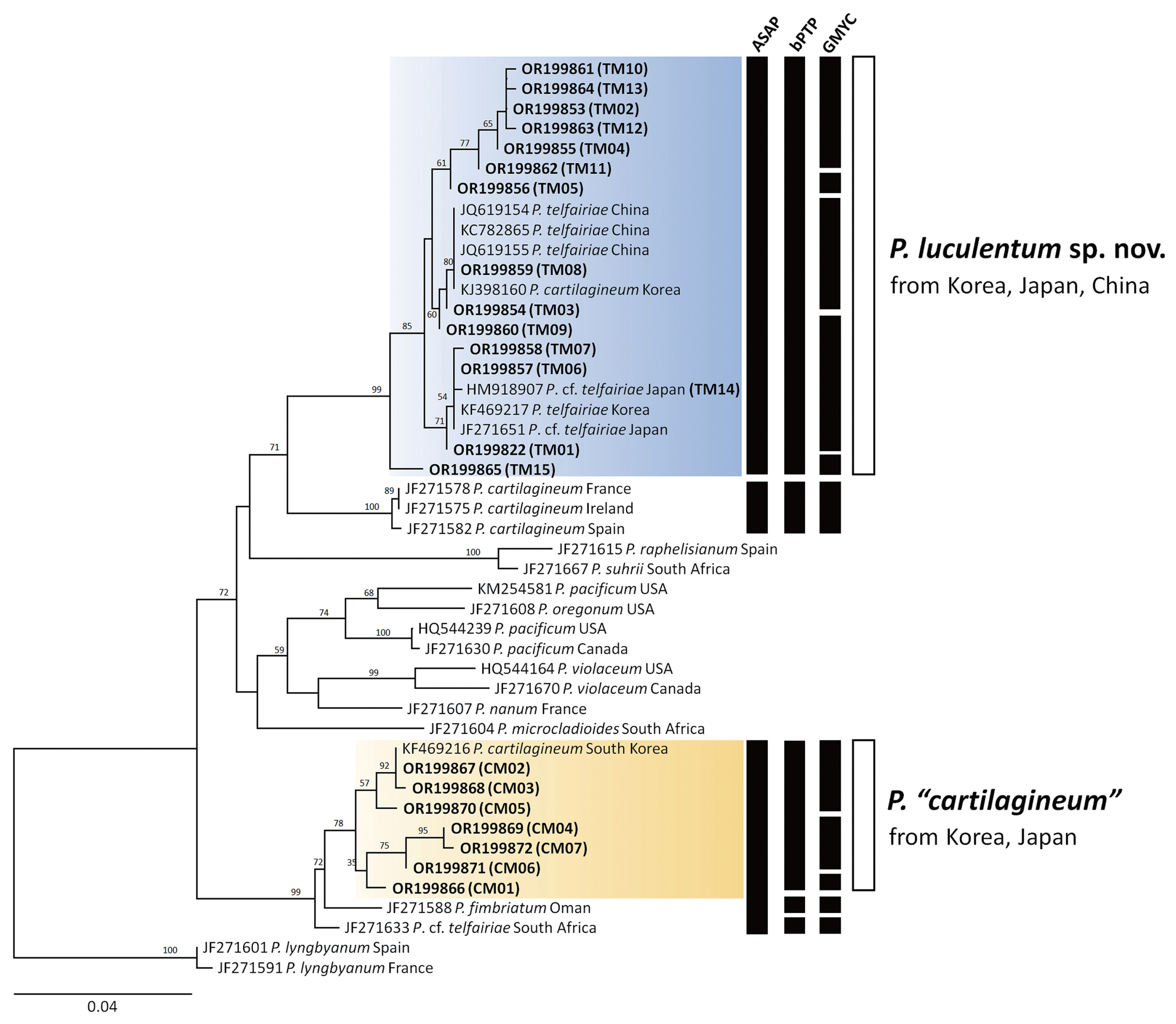
The COI-5P and rbcL datasets contained no ambiguous positions and had a median length of 655 bp (range, 350–660 bp) for COI-5P and 1,200 bp for rbcL, analyzing 209 specimens for the two presumed species of Plocamium from the NW Pacific. Phylogenetic analyses based on the COI-5P revealed two well-supported clades, identified as P. “telfairiae” and P. “cartilagineum” from the NW Pacific (Fig. 1). The P. “telfairiae” clade included 15 haplotypes (TM01–TM15) and clustered with GenBank sequences from Korea, Japan, and China. The NW Pacific P. “telfairiae” clade was sister to P. cartilagineum from Europe (JF271575, JF271578, and JF271582), which was confirmed as authentic P. cartilagineum. Another clade, P. “cartilagineum” from Korea, included seven haplotypes (CM01–CM07) with a GenBank sequence from Korea and was sister to P. fimbriatum from Oman (JF271588) and South African P. cf. telfairiae (JF271633). In the rbcL phylogenetic tree (Supplementary Fig. S1), the NW Pacific P. “telfairiae” clade included seven haplotypes (TR01–TR07) with two GenBank sequences. The NW Pacific P. “cartilagineum” clade included five haplotypes (CR01–CR05) with a GenBank sequence. The maximum intraspecific divergences based on COI-5P gene were 2.1% for P. “telfairiae” and 2.0% for P. “cartilagineum.” The interspecific divergences among two Plocamium species from the NW Pacific ranged from 6.3 to 8.3%. The NW Pacific P. “telfairiae” has 4.06–5.27% COI-5P divergence with the European P. cartilagineum and 5.72–7.07% with P. telfairiae from South Africa.
Species delimitation
The molecular species delimitation methods (ASAP, bPTP, and GMYC) did not produce congruent results for the NW Pacific species (Fig. 2). In the COI-5P result, the best ASAP score result grouped all P. “telfairiae” sequences into a single clade, whereas P. “cartilagineum” was lumped with P. fimbriatum (JF271588) and P. cf. telfairiae (JF271633) into a single entity. The bPTP method produced similar results as ASAP for P. “telfairiae”, accordingly we propose a new species below. However, the bPTP method delineated three groups as P. “cartilagineum”, P. fimbriatum, and P. cf. telfairiae. GMYC divided P. “telfairiae” into five groups and P. “cartilagineum” into three. In the rbcL result, the best ASAP score and bPTP method identified P. “telfairiae” as a single entity, whereas GMYC divided it into four groups (Supplementary Fig. S1). The bPTP and GMYC methods produced congruent results in P. “cartilagineum” as a single clade, separating P. fimbriatum from Oman as shown in the COI-5P phylogeny (Supplementary Fig. S1). Overall, ASAP provided conservative estimates of the putative species, whereas GMYC assignments indicated potential over-splitting.
Taxonomic treatment
A new species, Plocamium luculentum sp. nov. is proposed based on the misidentified P. “telfairiae” from the NW Pacific. However, the taxonomic treatment for P. “cartilagineum” from the NW Pacific remains unresolved because of the inconsistent results of the three molecular species delimitation methods.
Plocamium luculentum sp. nov. M. Y. Yang & M. S. Kim
Description
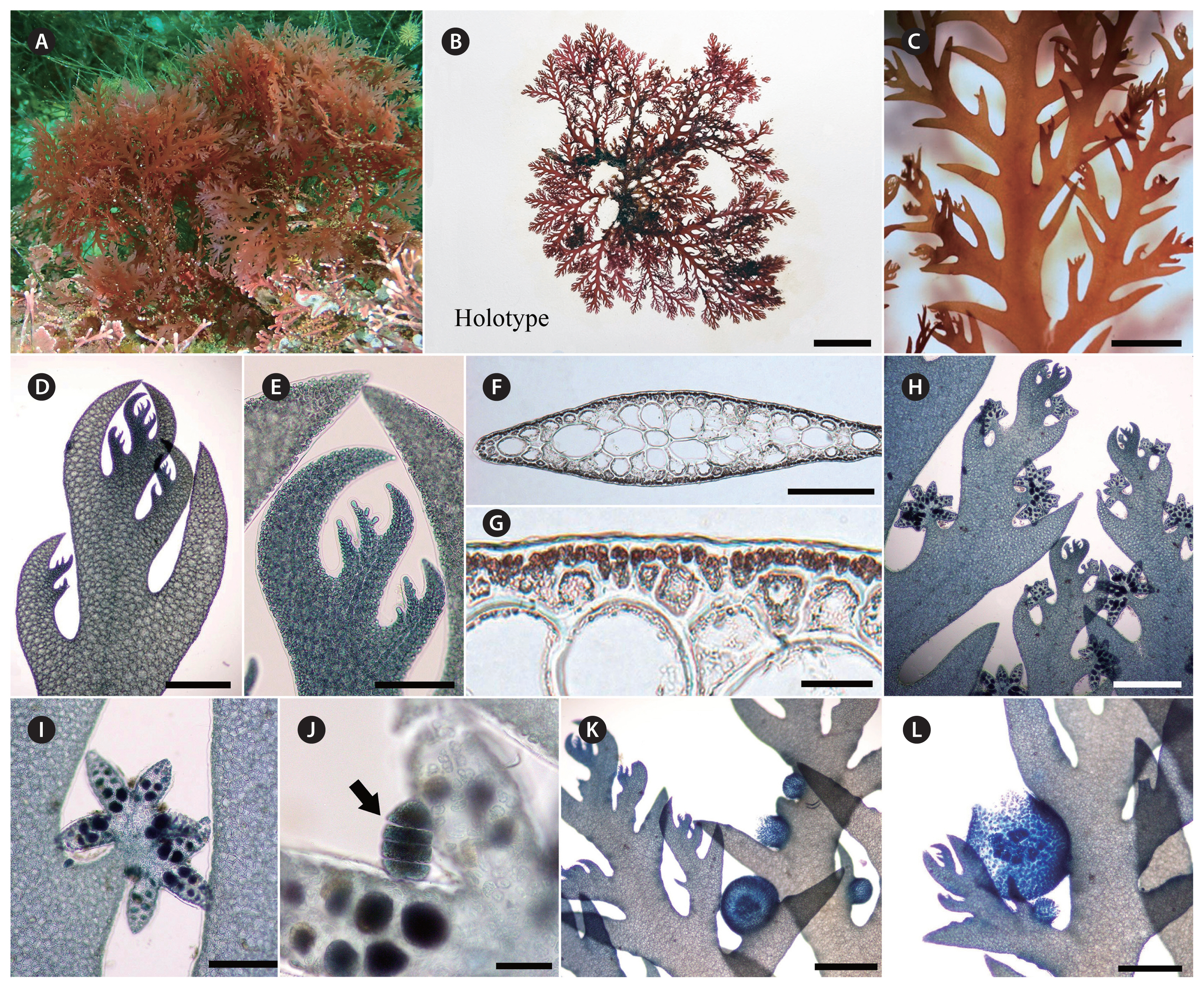
Thalli erect, flabellate, 7–20 cm in height, bright to dark red, densely branched, attached by a conspicuous holdfast (Fig. 3A & B); main axis 1.2–2.5 mm wide, thin membranous, complanate, 85–100 μm thick, branches developing in alternating pairs of 2–3 ramuli from margins of the axes (Fig. 3C–F); the lower pair of ramulus is usually determinate in growth, secund, spinose with acute tips (Fig. 3D–F); the upper pair of ramulus is usually well developed and has the potential to continue indeterminate in growth (Fig. 3D–F). Cortex consists of a layer of small pigmented cortical cells and 2–3 layers of large hyaline medullary cells (Fig. 3F & G). Tetrasporangial stichidia distinct and compact clustered, star-like, 440–490 μm in width, 250–310 μm in length, cylindrical or branched divaricately, scattered among the branches, and replacing ramuli in the series (Fig. 3H & I). Tetrasporangia divided zonately, 18–30 μm (Fig. 3J). Cystocarps sessile, globular on the margins of branches (Fig. 3K & L).
Holotype
JNUB–PT028, vegetative plant, collected on Jan 20, 2015, deposited in the herbarium of the JNUB. GenBank accession No. COI-5P: OR199853, rbcL: OR199875.
Genetic diversity, haplotype network, and distribution
In the COI-5P data set, the haplotype and nucleotide diversity were higher in P. luculentum sp. nov. (H = 0.8096 ± 0.0225 and π = 0.0105 ± 0.0056) compared with P. “cartilagineum” (H = 0.6583 ± 0.0279 and π = 0.0009 ± 0.0053) (Table 1).
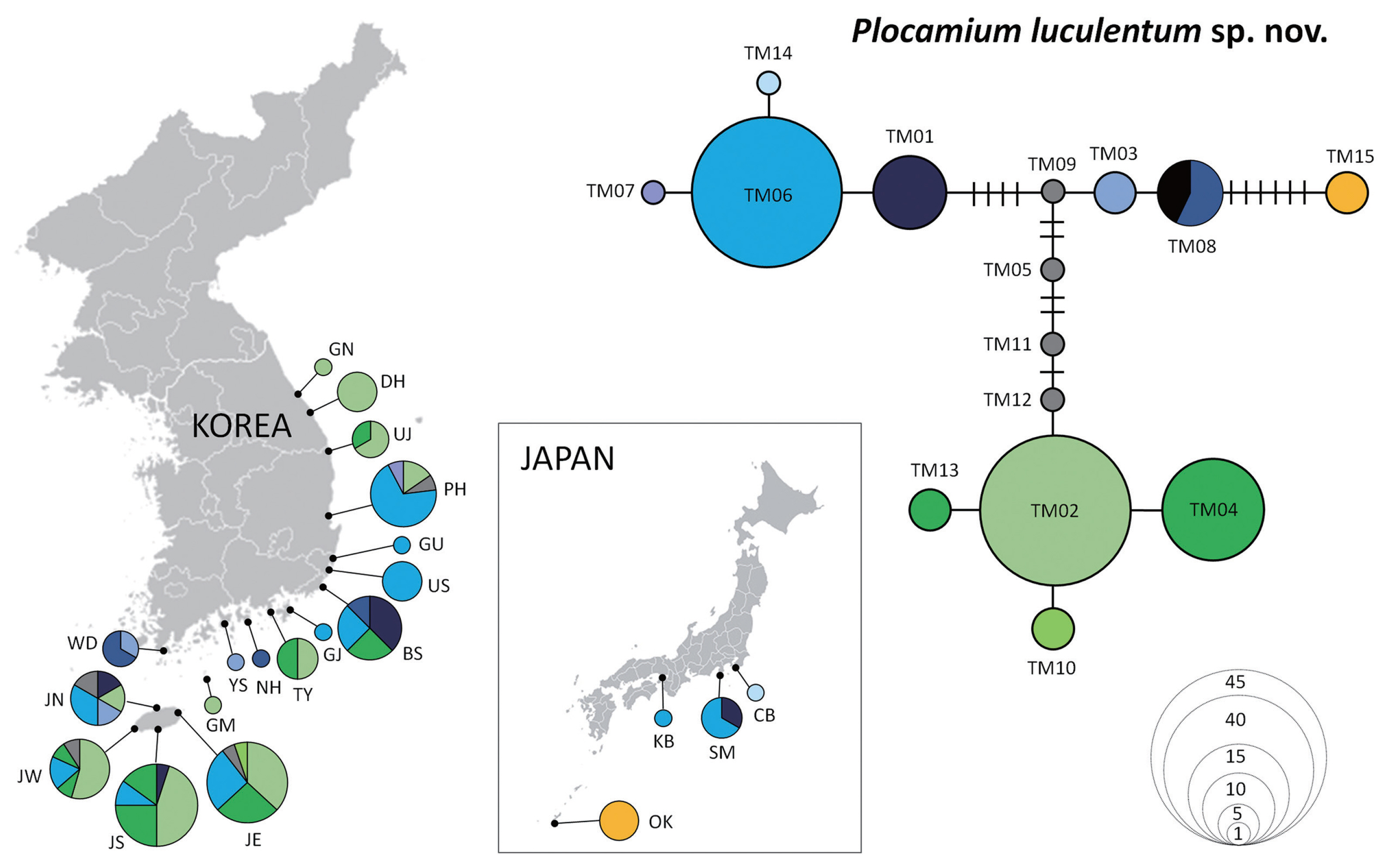
The haplotype network of P. luculentum sp. nov. recovered 15 haplotypes (Fig. 4, Supplementary Table S1), with two dominant haplotypes: TM02 (32.1%) and TM06 (28.4%). Haplotype TM02 was distributed in Korea only, covering areas from the south (Southern Jeju [JS]) to the north (Gangneung [GN]), whereas haplotype TM06 was found in Korea and Japan. Haplotype TM08 was shared among individuals from southern Korea (Wando [WD], Namhae [NH], and Busan [BS]) and China. Haplotype TM15 differed from TM08 in seven mutational steps and was found only in Okinawa, Japan.
For P. “cartilagineum”, seven haplotypes were detected (Fig. 5, Supplementary Table S2) with two dominant haplotypes, CM04 (45%) and CM02 (36%), which were distributed along the Korean coast. Another haplotype, CM01, was found in southern Korea, not above BS (Fig. 5). Haplotype CM05 differed from CM02 by four mutational steps and was found only in Okinawa, similar to the haplotype TM15 of P. luculentum.
Pairwise FST values based on COI-5P data (Table 2) revealed low genetic differentiation in P. luculentum from Korea (FST = 0.1386–0.1622).
DISCUSSION
Molecular-based species delimitation methods often result in the detection of more diversity than those previously identified based on morphology only. This study confirmed the presence of cryptic diversity among species of Plocamium previously identified as P. “telfairiae” and P. “cartilagineum” in Korea, Japan, and China. The findings of this study represent evidence of undescribed species in the genus Plocamium in the NW Pacific region (Kim and Hwang 2015) and may provide useful information where accurate species identification is needed for potential benefits, such as pharmaceuticals and food (Shilling et al. 2019, 2021, Park et al. 2022). Molecular analyses revealed significant differences between specimens of the two Plocamium species in the NW Pacific and those from their type localities. At least two endemic cryptic entities of Plocamium were found to occur in sympatry in the NW Pacific similar to that reported in Europe (Cremades et al. 2011). Previous studies have demonstrated a high level of biodiversity and endemism in marine macroalgal flora in the NW Pacific (Yang and Kim 2018, Kang et al. 2021). According to these results, DNA barcoding using the COI-5P mitochondrial marker highlighted genetic variation of endemic species with haplotype distribution from Korea and Japan. The genetic variation of the two cryptic entities from the NW Pacific exhibited much higher COI-5P divergence than the average genetic variation found within South African Plocamium species (Reddy et al. 2023). The maximum uncorrected divergence of COI-5P within these taxa was 2.1%, and 15 mitotypes in 109 specimens of P. luculentum sp. nov. and 7 mitotypes in P. “cartilagineum” with 2.0% were detected in 100 specimens. Contrary to the results of this study, the intraspecific variation of the three recently reported Chilean Plocamium species were 0.02, 0.04, and 0.14%, respectively (Montecinos et al. 2021). Moreover, morphological variation among Plocamium species have been observed worldwide (Yano et al. 2004). Therefore, the two cryptic entities detected in this study represent a complex of poorly understood species rather than being identical to genuine species (Cremades et al. 2011). Based on these results, we report a new species, P. luculentum sp. nov. that was previously misidentified as P. “telfairiae” from Korea. However, regarding Korean P. “cartilagineum”, the report of a new species is postponed owing to the incongruent results of molecular-based species delimitation methods. Investigations involving more samples from the type locality or type specimens of P. telfairiae and detailed morphological observation may clarify the uncertainty of P. “cartilagineum” from the NW Pacific in the future.
The type locality of P. telfairiae is Mauritius in the Indian Ocean, off the eastern coast of Africa (Guiry and Guiry 2023). In this study, P. luculentum sp. nov., previously misidentified as P. “telfairiae” in Korea, Japan, and China, was phylogenetically distant to the South African P. telfairiae (JF271633) (Montecinos et al. 2021, Reddy et al. 2023). The interspecific divergence of COI-5P data analysis ranged from 5.72–7.07%. The reconstructed phylogenetic tree showed that P. luculentum sp. nov. formed a sister group with the European P. cartilagineum clade (Cremades et al. 2011), and their interspecific variation was 4.06–5.27%. Plocamium luculentum sp. nov. represents the most widely distributed species of Plocamium in Korea and typically grows in subtidal areas and as an epiphyte on other algae (Kim and Hwang 2015). The specimens were collected from the south to the east coasts of Korea and P. “telfairiae” has been reported in most areas in Japan (Yano et al. 2004, 2005), as well molecularly confirmed in China (Cremades et al. 2011). Yano et al. (2004, 2005) reported that Japanese P. “telfairiae” is not clearly distinguished from Japanese P. “cartilagineum” based on diagnostic characteristics, suggesting that physiological properties such as intracellular pH may be informative to understanding species delimitation owing to morphological homoplasy among Japanese Plocamium species. Comparing the conventional diagnostic traits of Korean and Japanese specimens of P. “telfairiae”, there were no significant differences in the size of the lowermost branchlet and the main axes, and the number of branchlets in each alternate series mostly having two branchlets. Therefore, at least two Japanese (JF271651 and HM918907) and three Chinese specimens (JQ619154, JQ619155, and KC782865) (Cremades et al. 2011) included in the phylogenetic tree may belong to the same taxon as the new species, P. luculentum sp. nov.
Saunders and Lehmkuhl (2005) indicated that P. cartilagineum has substantial genetic diversity among specimens from different geographic locations that share some morphological similarities and that Japanese P. cartilagineum differed from representatives of this species in northern Europe based on LSU sequences (Saunders and Lehmkuhl 2005). Korean P. “cartilagineum” was confirmed as a different species because it was not resolved as sister to the true P. cartilagineum from Europe (JF271575, JF271578, and JF271582) (Cremades et al. 2011). The interspecific variation of Korean P. “cartilagineum” and genuine European P. cartilagineum specimens is 6.03–7.46%. Lee and Kim (2022) revealed that Korean P. “cartilagineum” grows on rocky substrate in the intertidal zone with extreme morphological variation and reported differences in the morphology of tetrasporangial stichidia, size of cystocarps, and width of lower branchlets between Korea and European specimens (Lee and Kim 2022). Yano et al. (2004) demonstrated that Japanese species have considerable morphological variability, which is linked to water motion and nutrient availability, as confirmed in a study on Korean species (Lee and Kim 2022).
In the COI-5P phylogenetic tree (Fig. 1), the Korean P. “cartilagineum” specimens grouped with P. fimbriatum from Oman (JF271588) and South African P. cf. telfairiae (JF271633) (Cremades et al. 2011). Their genetic variation was 1.81–2.72% for Korean P. “cartilagineum” and P. fimbriatum from Oman and 1.63–2.36% for Korean P. “cartilagineum” and South African P. cf. telfairiae. Although the three entities are genetically similar and form a monophyletic clade with strong support, they have different morphological characteristics, as well as distribution areas (De Clerck et al. 2002, Wynne 2002, Reddy et al. 2023). Thus, the possibility of a cryptic species in the Korean specimens cannot be excluded. Furthermore, molecular-based species delimitation revealed different results depending on the method used (ASAP, bPTP, or GMYC). bPTP indicated the existence of at least three species, including Korean P. “carilagineum”, P. fimbriatum from Oman, and South African P. cf. telfairiae. Therefore, further work is required to resolve the taxonomy of this clade, including additional DNA sequences from the Indian Ocean. P. fimbriatum from Oman was described as a new species by Wynne (2002), distinguished by robust main axes 25 cm in length and 4–6 mm in width, abundant marginal proliferation, and tetrasporangial stichidia arising along the axillary margins and in the proliferation. Despite having phenotypic plasticity, comparing the morphology of Korean P. “cartilagineum” and P. fimbriatum from Oman revealed that the Korean species is 5.5 cm in length and 0.6 mm in width, without marginal proliferations, and has tetrasporangial stichidia arising on the upper margin of ramuli (Lee and Kim 2022). In addition, P. fimbriatum has laterals in alternating pairs; however, the ramuli of the Korean species are in an alternating series of 2–3(4). Consequently, the morphology of P. fimbriatum differed from that of Korean P. “cartilagineum” in robustness and height, extensive prostrate system, and broad primary axes. In addition, the thalli of South African P. telfairiae are 9 cm high and 1(−2) mm wide in the main axes, with ramuli forming in alternate-distichous series of one simple and one compound branch, and tetrasporangial stichidia in the axils of ramuli or replacing a compound ramulus (De Clerck et al. 2002). Although these features of the South African specimens agree with descriptions and isotype herbarium specimens in BM, the Korean P. “cartilagineum” has ramuli in alternating series of 2–3, unlike the paired arrangement in South African P. telfairiae (De Clerck et al. 2002).
This study is the first intensive phylogeographical study of two Plocamium species in Korea, showing a lack of significant correlation between genetic differences and geographic distribution. Moreover, in the haplotype network analysis, major haplotypes, TM02 and TM06, were detected for P. luculentum sp. nov., and CM02 and CM04 for P. “cartilagineum.” These major haplotypes in both species existed between distant populations along the Korean coast, indicating that the dispersal of these species may occur over long distances in the NW Pacific. The relationships of haplotypes did not confirm the geographic distribution between samples, suggesting that the genetic structure of P. luculentum sp. nov. is weak (Wei et al. 2023).
Low genetic differentiation, as observed in both Plocamium species in this study, has also been observed in other marine macroalgae in the NW Pacific: Pachymeniopsis elliptica / P. lanceolata (Yang et al. 2021b), Chondrus ocellatus / C. nipponicus (Yang and Kim 2022), and Gratelopia asiatica / G. jejuensis (Yang et al. 2021c). The absence of distinct geographic genetic structures reflects an apparent lack of major oceanographic barriers in Korea. The combination of northward (East Korea Warm Current, a branch of the Kuroshio Current) and southward (North Korea Cold Current) oceanographic flows may allow effective population mixing. The analysis of genetic diversity based on the COI-5P showed high haplotype diversity and low nucleotide diversity for P. luculentum and P. “cartilagineum.” This result is consistent with a rapid population expansion event over historical time intervals, where populations do not accumulate large nucleotide variation because of small effective population sizes (Grant and Bowen 1998 ).
Although genetic connectivity was demonstrated in the NW Pacific, regional haplotypes were present, suggesting a unique phylogeographic pattern. Haplotypes distributed only in Okinawa, such as TM15 for P. luculentum sp. nov. and CM05 for P. “cartilagineum”, did not overlap with other populations indicating the need for further screening to detect other regional haplotypes. The unique haplotype and geographical distribution can inform us on historical refugia (Chiocchio et al. 2021), hotspot regions with high genetic diversity (Yang et al. 2021a), and conservation planning. Genetic hotspots were found in southern Korea for P. luculentum sp. nov., and Jeju for P. “cartilagineum.” Low genetic diversity was found in eastern Korea for both species, particularly for P. “cartilagineum.” The haplotype distribution map showed simple genetic structure in Uljin (UJ), Donghae (DH), and Gangneung (GN) for P. luculentum sp. nov. and in Donghae (DH) and Yangyang (YY) for P. “cartilagineum” (Figs 4 & 5), suggesting that these two species may lost their genetic diversity during recolonization to the north.
The widely distributed taxa such as several red algal species may have areas of sparse genetic diversity, making these populations susceptible to extinction (Nauer et al. 2019, Díaz-Tapia et al. 2020, Boo et al. 2022). Therefore, recognizing hotspots of intraspecific genetic diversity could aid biodiversity conservation (Yang et al. 2020, Yang and Kim 2022). Effective conservation of taxa requires accurate knowledge of genetic variation within species. This study highlights the need for more research on macroalgal cryptic diversity in the NW Pacific, as well as a global revision of the diversity and biogeography of Plocamium.